Algorithms capable of detecting large-scale chromosomal abnormalities with a high degree of accuracy developed
Could contribute to the investigation into the cause of cell cancerization and early detection of cancers
A group of researchers led by SESE Jun (Chief Researcher, Computational Functional Genomics Research Group, Biotechnology Research Institute for Drug Discovery, National Institute of Advanced Industrial Science and Technology (AIST)), YAMAGATA Koichi (former special researcher, AIST, currently Assistant Professor, Chuo University), and TAKEDA Junji (Professor, Graduate School of Medicine, Osaka University) has developed software called "COntrol SaMple-based detectiOn of Structural variation (COSMOS)," which includes algorithms capable of detecting large-scale chromosomal abnormalities (structural variations) with a high degree of accuracy and sensitivity.
In recent years, structural variations characteristic of cancerized cells have been frequently observed. Detecting such structural variations is expected to lead to early detection of cell cancerization and the selection of appropriate treatment methods based on the analysis of structural variations.
COSMOS is software for analyzing big data of genome sequences obtained by super parallel sequencing. This group conducted validation of COSMOS by using mice embryonic stem cells and it achieved higher accuracy and sensitivity than that of other methods.
This group's technology may contribute to the investigation into the cause of cell cancerization and early detection of cancers.
Abstract
An important challenge in cancer genomics is precise detection of structural variations (SVs) by high-throughput short-read sequencing, which is hampered by the high false discovery rates of existing analysis tools. Here, we propose an accurate SV detection method named COSMOS, which compares the statistics of the mapped read pairs in tumor samples with isogenic normal control samples in a distinct asymmetric manner. COSMOS also prioritizes the candidate SVs using strand-specific read-depth information. Performance tests on modeled tumor genomes revealed that COSMOS outperformed existing methods in terms of F-measure. We also applied COSMOS to an experimental mouse cell-based model, in which SVs were induced by genome engineering and gamma-ray irradiation, followed by polymerase chain reaction-based confirmation. The precision of COSMOS was 84.5%, while the next best existing method was 70.4%. Moreover, the sensitivity of COSMOS was the highest, indicating that COSMOS has great potential for cancer genome analysis.


Figure 1
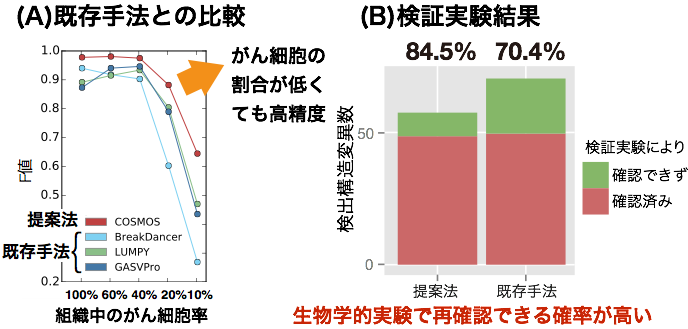
Figure 2
To learn more about this research, please view the full research report entitled " COSMOS: accurate detection of somatic structural variations through asymmetric comparison between tumor and normal samples " at this page of the Nucleic Acids Research website.
Related links
