Genomic cut and paste using a Class 1 CRISPR system
Japanese researchers led by Osaka University show that a Class 1 CRISPR gene editing system can achieve functional DNA repairs in human cells with no prominent off-target effects
Almost from the moment DNA was discovered, the ability to fix or remove disease-causing genes in affected patients has been something of a holy grail of medicine. Now that this goal is within reach, researchers are working to fine-tune the technology to ensure safe and effective gene editing with no unwanted downstream effects.
In a study published this month in Nature Communications , Japanese researchers led by Osaka University describe a new genome editing approach that brings us much closer to achieving the dream of eliminating a host of diseases caused by faults in our DNA.
Most modern gene editing techniques rely on CRISPR technology—short for “clusters of regularly interspaced short palindromic repeats”—adapted from bacterial anti-viral defense systems. Essentially, CRISPR-associated proteins (Cas) can be directed to specific regions of the genome, where they make precise cuts in the DNA. This approach can be used to completely eliminate regions of faulty DNA or to introduce new DNA by supplying an altered blueprint that is followed by the cell when repairing the DNA break.
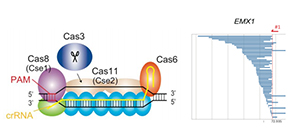
There are two main classes of CRISPR systems, Class 1 and Class 2, that are distinguished by the number of helper proteins needed to cut the DNA. While most gene editing approaches use single-component Class 2 CRISPR systems, very little is known about the utility of multi-component Class 1 systems in eukaryotic gene editing.
“Class 2 CRISPR systems, particularly those using the Cas9 or Cas12a enzymes, are widely used for eukaryotic genome editing,” says lead author of the study Hiroyuki Morisaka. “However, these systems aren’t perfect: as well as introducing unintended mutations, genome editing efficiencies using these methods can be somewhat variable.”
The researchers therefore decided to investigate whether Class 1 CRISPR systems could offer a more efficient and safer alternative.
Using a Cas3 protein-based Class 1 CRISPR system, the team successfully demonstrated both DNA deletions and insertions in human cells. Notably, the Cas3 protein induced unidirectional deletions of large sections of DNA, setting it apart from Class 2 enzymes which usually need help to achieve such large genome edits. Most importantly though, Cas3 achieved more efficient genome editing than Cas9, with no prominent off-target effects.
To confirm the therapeutic potential of the system, the researchers carried out Cas3-based repair of the DMD gene in induced pluripotent stem cells from a patient with Duchenne muscular dystrophy.
“Our results suggest that this Cas3-based method offers a superior alternative to Class 2 CRISPR gene editing systems,” says senior author Tomoji Mashimo. “As well as its obvious therapeutic uses, we envisage potential applications in drug discovery, disease prevention, and crop improvement.”
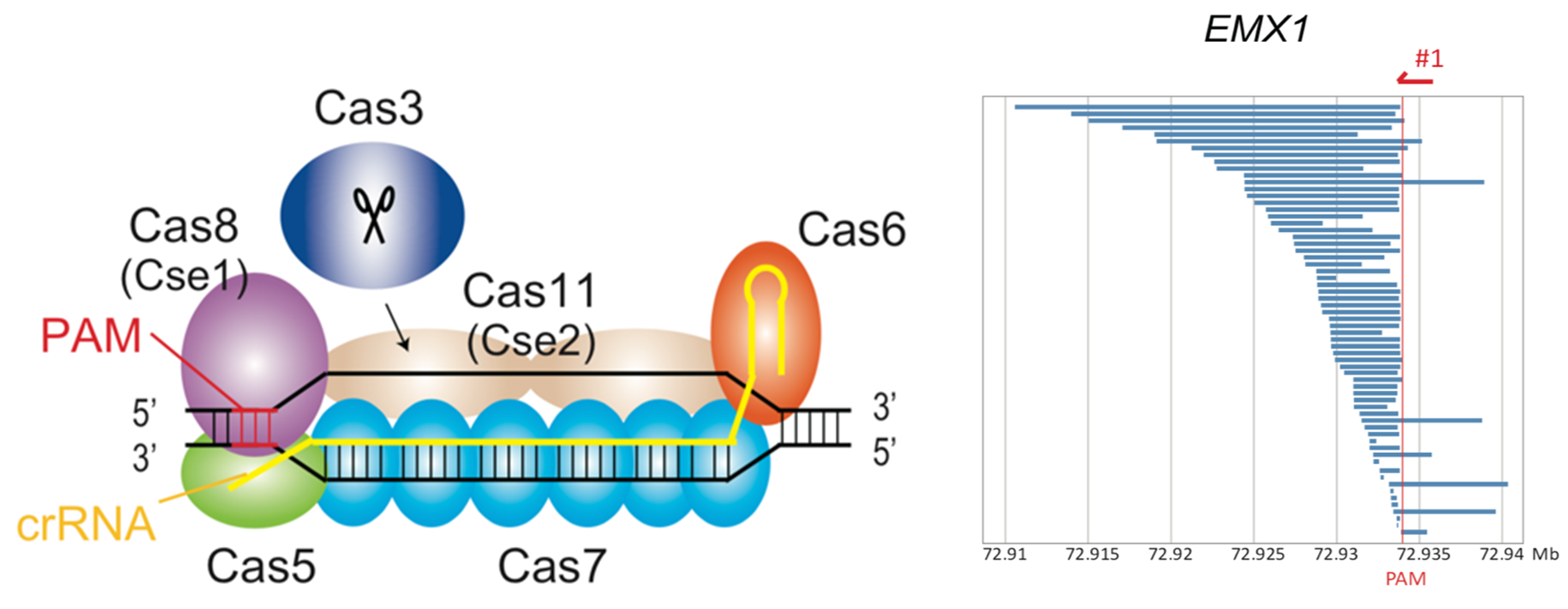
Fig. 1. CRISPR-Cas3 system mediates DNA cleavage in human cells. (Left) Type I-E CRISPR effector is composed of crRNA, Cas3, and a large Cascade complex, which contains Cas5, Cas6, multiple Cas7, Cas8 recognizing the PAM, and two Cas11. (Right) Cas3-mediated DNA deletion patterns via microarray-based capture sequencing at EMX1 and CCR5 loci in 293T cells.

Fig. 2. Cas3-mediated DMD exon skipping in induced pluripotent stem cells. (Left) RT-PCR analysis of the DMD exon 45 skipping in skeletal muscle cells differentiated from subclones of DMD patient-derived iPSCs treated with Cas3. (Right) The restored DMD protein detected in skeletal muscle cells differentiated from the exon skipping subclones of DMD-iPSCs.
The article, “CRISPR-Cas3 induces broad and unidirectional genome editing in human cells,” was published in Nature Communications at DOI: https://doi.org/10.1038/s41467-019-13226-x .
Related links
