Systemic lupus erythematosus linked to altered gut microbiome
Researchers from Osaka University reveal changes in the components of the gut microbiome that are associated with the autoimmune disease systemic lupus erythematosis
Systemic lupus erythematosis (SLE) is an autoimmune disease in which the immune system targets tissues of the body, causing widespread inflammation and affecting multiple organs such as the kidney and the brain. The gut “microbiome”—all the micro-organisms that live in the human gut—is known to be altered in SLE patients. Now, a research team at Osaka University has comprehensively profiled the associations between the gut microbiome and SLE.
Healthy human intestines contain billions of micro-organisms that are essential for the normal functioning of the gut. They protect against pathogens, assist in the metabolism of food, and are also able to affect the immune response. Various diseases have been linked to disturbances of the gut bacteria leading to imbalance, or “dysbiosis,” including autoimmune conditions.
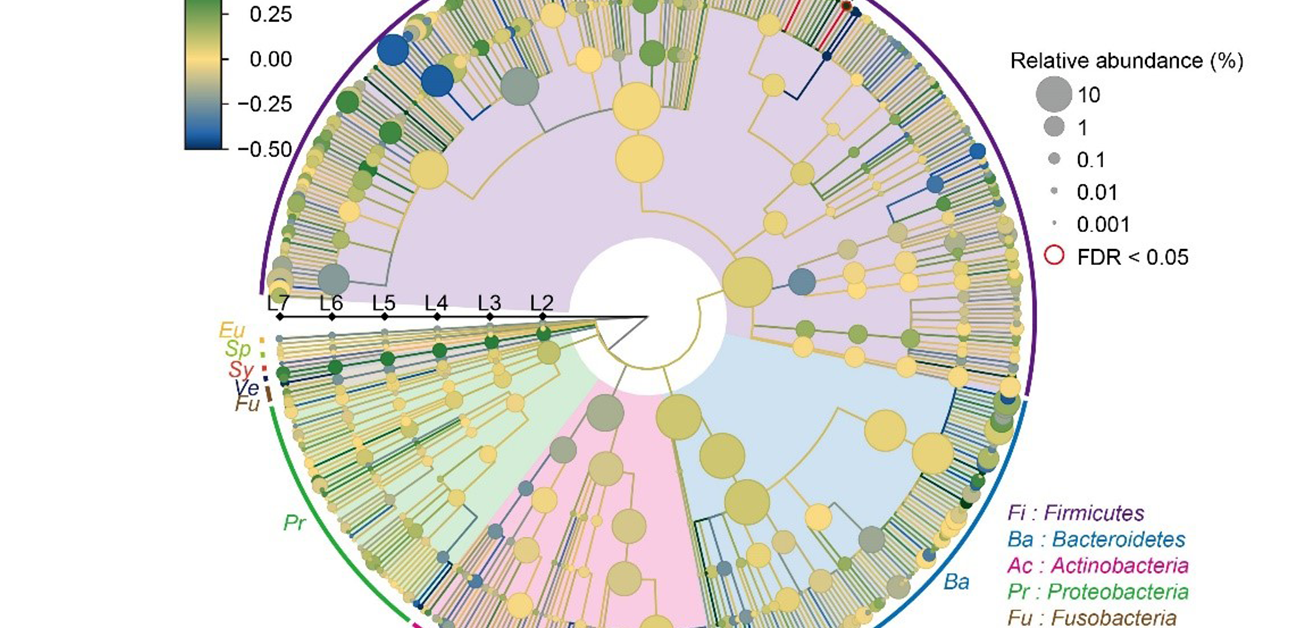
The team isolated DNA from the intestinal microbiome using fecal samples, and then used a next-generation sequencing machine to carry out metagenome shotgun sequencing. This is a technique that allows the sequencing of all genes present in a complex sample by fragmenting all the genomes, sequencing the short fragments, and then reassembling the sequences. By collecting samples from both SLE patients and healthy comparisons, they could comprehensively assess the relationship between the gut microbiome and SLE. “We were able to show distinct changes to the gut microbiome in SLE patients,” says lead author Yoshihiko Tomofuji, “as we found that two species of Streptococcus bacteria, Streptococcus anginosus and Streptococcus intermedius, were significantly increased in the gut microbiome of patients with SLE.”
The gut microbiome can affect the wider functions of the body by altering the population of small molecules, or metabolites, circulating in the liquid part of the blood, called the plasma. The team therefore went on to integrate the gut microbiome data with data on the entire population of plasma metabolites, known as the plasma metabolome.
“Our analysis revealed interactions between the microbiome and the host that were mediated by the metabolome,” explains senior author Yukinori Okada. “A particular molecule known as acylcarnitine had a positive correlation with the SLE-associated bacterium.”
Acylcarnitine is known to induce inflammation and could therefore potentially act as a trigger for the overactivation of the immune system seen in SLE.
This research reveals for the first time the specific microbial landscape of SLE patients, contributing to our understanding of the relationship between the gut microbiome and SLE and providing useful resources for future research.
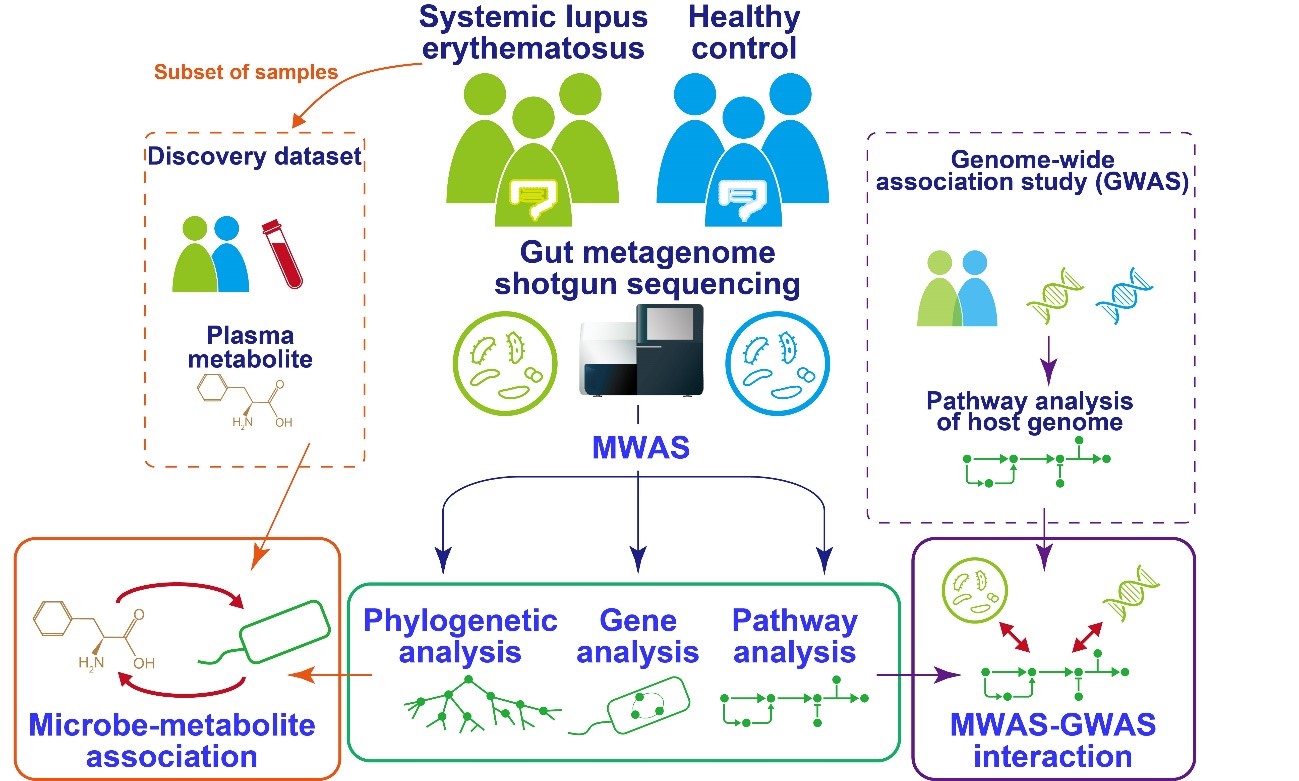
Fig.1 Overview of the study.
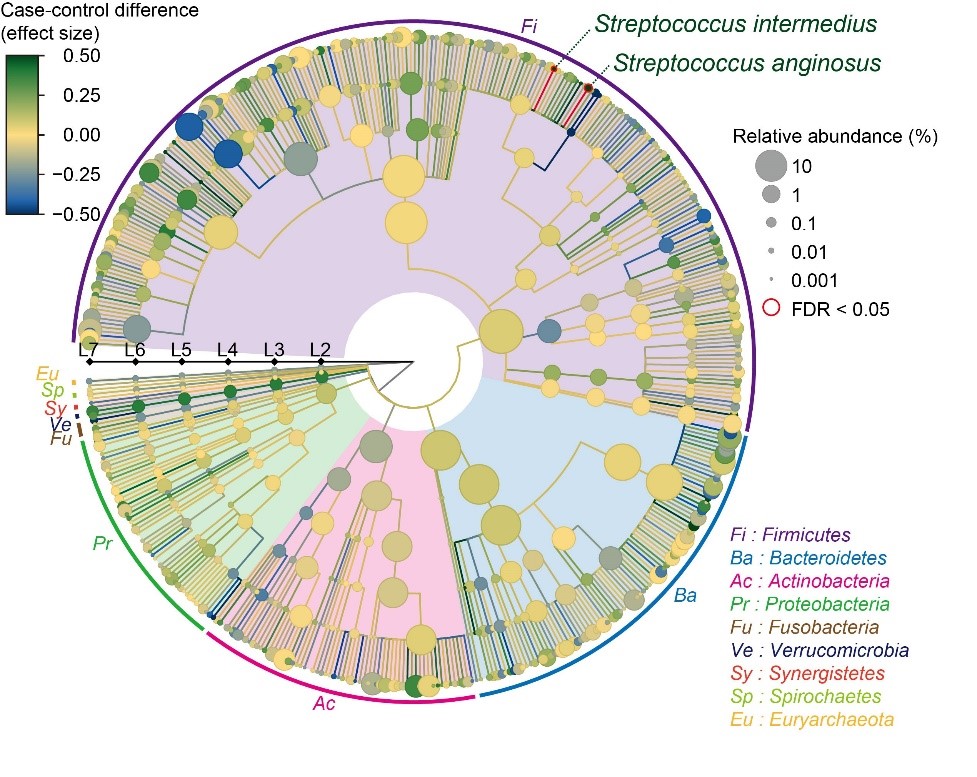
Fig.2 Result of the case–control comparison of the microbial taxa is indicated as a phylogenetic tree. Streptococcus anginosus and Streptococcus intermedius increased in the SLE patients, with a false discovery ratio < 0.05. (credit: 2021 Yoshihiko Tomofuji et al., Annals of the Rheumatic Diseases)
The article, “Metagenome-wide association study revealed disease-specific landscape of the gut microbiome of systemic lupus erythematosus in Japanese,” was published in Annals of the Rheumatic Diseases at DOI: https://doi.org/10.1136/annrheumdis-2021-220687.
