Traits and specific regions of the human genome related to human adaptive evolution identified: Drinking for Japanese and bread for Westerners
A group of researchers led by Professor OKADA Yukinori from Osaka University identified 29 genetic loci with selection signatures in the Japanese population, using the ascertained sequentially Markovian coalescent (ASMC), from the biobank-based large-scale genome-wide association study data of 170,882 subjects. The researchers also revealed that selection signatures in evolution were different between the Japanese population and the European population by analyzing data from the UK Biobank resource.
Adaptive evolution in humans is promoted by evolutionary processes like natural selection due to their geographical condition and living environment. This research group previously identified 4 genetic loci related to selection signatures by conducting a genome-wide scan of 2,000 Japanese individuals, reporting that alcohol metabolism was related to selection signatures in the Japanese population. (Okada Y et al. Nature Communications 2018)
This time, the group conducted a genome-wide scan of selection signatures of the Japanese population from the biobank-based large-scale genome-wide association study data of 170,882 subjects.
Using ASMC, which can estimate the locus-specific time to the most recent common ancestor, they identified 29 genetic loci with selection signatures satisfying the genome-wide significance within the past 20,000 years. As a result, they found significant positive selection pressure at the functional missense variant of ADH1B, which is associated with lower alcohol consumption.
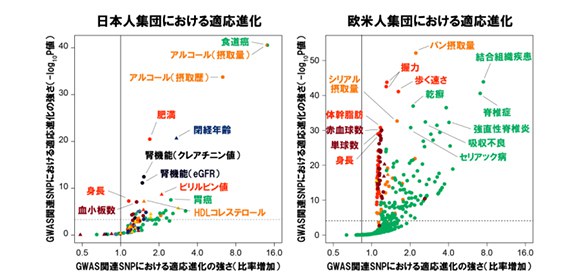
In addition, by examining the relationship between (a) genetic variants that affect more than 100 traits (such as onset of disorder, clinical test results, and dietary habits) in the Japanese population and (b) natural selection pressure, they identified selection enrichments in drinking-related behavior (such as alcohol drinking dose, alcohol drinking history) as well as kidney disorder, obesity, and immune-related diseases.
Then, they analyzed enrichment of more than 600 traits in the European population (110,000 individuals) using the UK Biobank resource, finding that the enrichment in anthropometric traits was observed for bread intake, hand grip strength, walking pace, and spondylopathy.
From these findings, it is thought that adaptive evolution in the population has been driven by natural selection and that dietary habits, such as alcohol intake and bread consumption, have also been deeply related to human adaptive evolution.
This group clarified specific regions of the human genome and traits related to adaptive evolution in the Japanese population, which will elucidate the history of adaptive evolution in the Japanese population. This study shows that utilizing biobank-scale genome data contributes to the clarification of common disease genomics and adaptive evolution of humans.
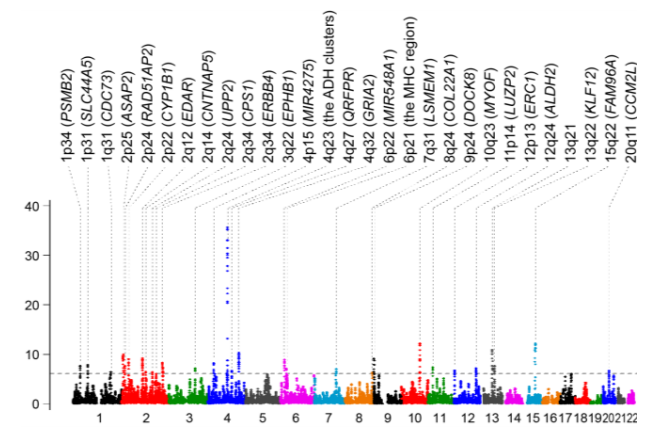
Figure 1
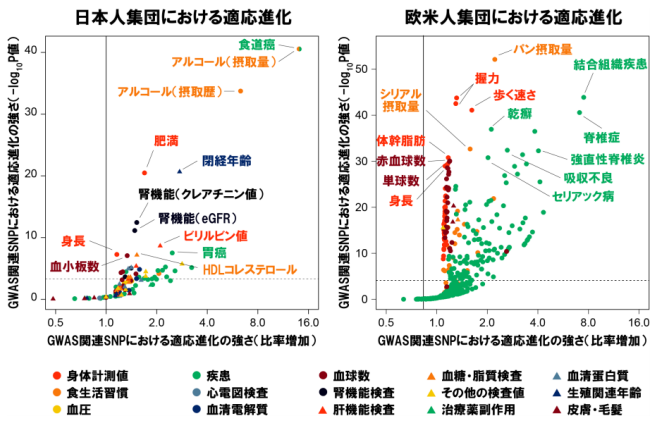
Figure 2
The article, “Genome-wide natural selection signatures are linked to genetic risk of modern phenotypes in the Japanese population,” was published in Molecular Biology and Evolution at DOI: https://doi.org/10.1093/molbev/msaa005.
