New susceptibility loci for type 2 diabetes in East Asian groups identified
A group of researchers from The University of Tokyo, the IMS RIKEN Center for Integrative Medical Sciences, and Osaka University conducted a meta-analysis of four Genome Wide Association Studies (GWAS) of about 400,000 cases, identifying 61 loci newly implicated in Type 2 diabetes (T2D) predisposition.
T2D is a common metabolic disease primarily caused by insufficient insulin production and/or secretion by the pancreatic β cells and insulin resistance in peripheral tissues. If T2D goes untreated, the high blood sugar can affect various organs and may lead to the development and progress of stroke, cardiac infarction, kidney failure, and cancer. It is said that 10 million people in Japan and more than 400 million people worldwide have T2D.
In East Asian populations, T2D prevalence is greater than in European populations among people of similar body mass index (BMI) or waist circumference; however, genetic factors in East Asians were not fully understood.
Base pairs of genetic letters in humans were almost identical in every person but individuals are 0.1 percent different genetically from every other person. Genetic information is inscribed in a linear molecule DNA, which is composed of four chemicals called bases. The order of the bases in the human genome can change, commonly at a single location. This kind of variation is called a single nucleotide polymorphism (SNP). SNPs are the most common type of genetic variation among people.
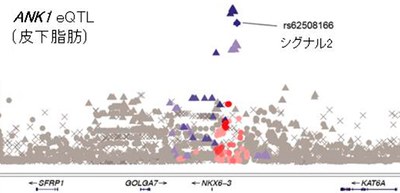
GWA using SNPs as genetic markers have successfully mapped thousands of loci associated with complex traits. In GWAS, genetic polymorphism, presence of two or more variant forms of a specific DNA sequence related to the development of diseases, are identified. However, since the identified SNPs are in regions of linkage disequilibrium that stretch over large distances, it is difficult to ascertain which gene in the region is functionally relevant.
Most of the GWAS signals that provide statistical evidence for increased risk of complex diseases map to non-coding regions. GWAS have identified several susceptibility loci for diseases, but the majority of identified SNPs fall within the non-coding region. The GWAS SNPs are more likely to be in genomic loci associated with gene expression as identified by expression quantitative trait loci (eQTL) mapping.
Thus, this group analyzed eQTL data and GWAS data and suggested that two independent genetic signals (or specific SNPs, which are usually attributed to the genes closest to the polymorphic markers that display the strongest statistical association) in different tissues might increase the risk of T2D through cis-eQTL variant.
They also found:
- T2D loci were also identified at clusters of noncoding RNAs with roles in islet β cell function. One locus included a set of microRNAs (miRNAs) specifically expressed in islet β cells.
- The other noncoding RNA locus was the MIR17HG cluster of miRNAs that regulate glucose-stimulated insulin secretion and pancreatic β cell proliferation stress.
- Another T2D locus was located near TRAF3, which is a direct target of the MIR17HG miRNA cluster and promotes hyperglycemia by increasing hepatic glucose production.
These findings suggested that in addition to genetic factors, a miRNA cluster and genes that affect muscle and adipose differentiation would influence the T2D susceptibility.
This study will deepen the understanding of hereditary factors of T2D in East Asians and lead to the clarification of T2D pathophysiology and the development of its therapeutic drugs.
Figure 1
The article, “Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population,” was published in Nature at DOI: https://www.nature.com/articles/s41588-018-0332-4.
See also:
Identification of type 2 diabetes loci in 433,540 East Asian individuals
Related Links
Department of Statistical Genetics, Graduate School of Medicine, Osaka University (link in Japanese)
