Mechanisms of stabilization of synthetic analogues of natural nucleic acids in helical structures clarified
A group of researchers from Nagoya University, Nagoya City University, the Explanatory Research Center on Life and Living Systems, and Osaka University has clarified the structural basis that allows Xeno nucleic acids (XNAs) to recognize natural nucleic acid RNA.
DNA and RNA are biological molecules consisting of phosphate group, sugar, and nucleobase and form stable duplexes with RNA in a sequence-specific manner. XNAs are formed of chains of linked nucleotides, similar to DNA and RNA but with a different sugar backbone, are expected to be useful in the development of nucleic acid drugs, nanotechnology applications, as well as a variety of approaches to study the origin of life.
Since nucleic acids are quickly broken down by nuclease enzymes, for designing nucleic acid drugs, it is expected to develop artificial or xeno nucleic acids that have high nuclease resistance and affinity with naturally occurring nucleic acids.
Previously, this group developed XNAs: Serinol nucleic acid (SNA) and acyclic (non-circular) L-threoninol nucleic acid (L-aTNA). Both of which have no sugar backbone and are derived from amino acids threonine and serine. The group also clarified that these acyclic nucleic acids were able to form stable duplexes with RNA in a sequence-specific manner.
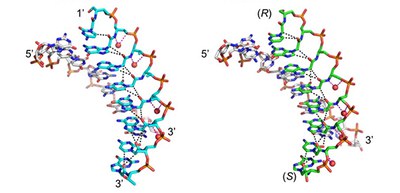
In this study, the group found that structures of RNA hybridizing with SNA and with L-aTNA were unwound right-handed helical structures just like natural nucleic acids but had large helical pitches. They also revealed that intrastrand hydrogen bonding networks between nucleobases and neighbouring backbones of L-aTNA and SNA enabled stabilization of acyclic structures and adjusted formation of heteroduplexes with RNA. The structural information the group gained will be helpful in designing guidelines for nucleic acid drugs that target RNA sequences related to diseases.
This group demonstrated that even acyclic artificial nucleic acids, in which amino acid derivatives are used as backbones, were able to form homoduplexes just like natural nucleic acids. These findings will also help to solve the mysteries of why DNA and RNA related to the origin of life have a sugar-phosphate "backbone" and why nature selected ribose as the backbone of genetic materials and amino acid as the backbone of proteins.
Figure 1
Figure 2
Figure 3
The article, “Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA,” was published in Communications Chemistry at DOI: https://www.nature.com/articles/s42004-020-00400-2.
Related Links
Uchiyama Laboratory, Graduate School of Engineering (link in Japanese)
