Cutting-edge analysis reveals how different drugs interact with the same target
Osaka University-led researchers use advanced technology to reveal the various ways different drugs bind to the same target, with potential implications for predicting therapeutic effects and optimizing drug design
Osaka University-led researchers use advanced technology to reveal the various ways different drugs bind to the same target, with potential implications for predicting therapeutic effects and optimizing drug design
---
Tumor necrosis factor (TNF) is involved in a range of inflammatory diseases including rheumatoid and psoriatic arthritis, ankylosing spondylitis, and psoriasis. Several drugs that target TNF are available to treat these conditions; however, despite acting on the same target, their clinical effectiveness and side effects differ.
Now, a team led by researchers at Osaka University has revealed differences in the sizes of drug-TNF complexes formed by three anti-TNF drugs, and the ratio of drug molecules to TNF molecules in these complexes. The researchers used a technique called sedimentation velocity analytical ultracentrifugation (SV AUC), which involves spinning a solution at very high speeds and investigating how quickly the centrifugal force causes molecules to move through the solution. From this information, the team could work out the size and shape of the TNF–drug complexes. The study was published in mAbs .
“Previous studies have investigated TNF binding with drug molecules,” lead author Elena Krayukhina says. “However, limitations of the techniques used have restricted researchers to looking at these interactions in very simple solutions, or with the molecules at relatively high concentrations. Using SV AUC and fluorescence detection, we were able for the first time to consider TNF–drug complex formation in the complex environment of human plasma, and at concentrations that reflect actual clinical conditions.”
The size of the complexes drugs form with their targets is important because it affects how the body responds to the drug. Research suggests that larger complexes are removed from the body more quickly than smaller ones. Larger complexes are also more likely to cause the body to launch an immune response against the drug, identifying and thus attacking it as an external threat. This can reduce the clinical effectiveness of the drug. The researchers found that one of the three drugs tested, etanercept, formed the smallest complexes with TNF and did not trigger a signaling pathway involved in immune response, unlike the other two drugs.
“These findings provide insight into possible mechanisms underlying observed differences in clinical efficacy and safety of these three anti-TNF drugs,” corresponding author Susumu Uchiyama says. “Our results also indicate that SV AUC is a valuable tool for investigating how drugs bind to their targets, which will help with predicting therapeutic action and optimizing pharmaceutical design in the future.”

Figure 1: Fluorescence-detected analytical ultracentrifugation analyses revealed that TNF-antagonists formed different complexes with TNF in human serum.
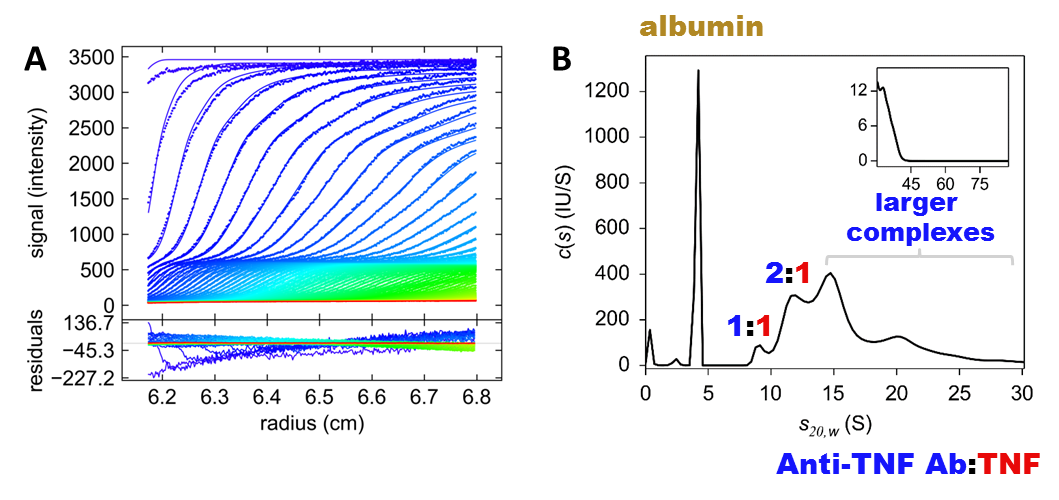
Figure 2: Representative raw fluorescence-detected analytical ultracentrifugation data acquired in human serum (A) and respective sedimentation coefficient distribution derived from data analysis (B).
To learn more about this research, please view the full research report entitled "Analytical ultracentrifugation with fluorescence detection system reveals differences in complex formation between recombinant human TNF and different biological TNF antagonists in various environments." at this page of mAbs.
Related link
Laboratory of Dynamic Cell Biology, Department of Biotechnology, Graduate School of Engineering, Osaka University
http://www.bio.eng.osaka-u.ac.jp/cl/english/
