How Protein Recognizes Invading Pathogens in Innate Immune Reaction Clarified
DNA binding clarified by this group will develop vaccine adjuvant and therapeutic drugs for viral infections and allergies
A group of researchers clarified the detailed steric constitution of TLR9 which detects invasion of microorganisms and activates innate immune reaction.
• Faculty of Pharmaceutical Sciences, The University of Tokyo -- SHIMIZU Toshiyuki (Professor), OHTO Umeharu (Lecturer), MIYAKE Kensuke (Professor), SHIBATA Takuma (Assistant Professor)
• Graduate School of Engineering, Osaka University -- UCHIYAMA Susumu (Associate Professor), KRAYUKHINA Elena (Specially Appointed Researcher)
As a system for preventing us from pathogen infection, an innate immune system and proteins named Toll-like receptors (TLR) are critical for our survival. TLR plays this role by being activated by molecules of pathogens and forming dimers.
Toll-like receptor 9 (TLR9) detects microbially-derived DNA sequences (CpG motifs) and urges the production of interfelon. TLR9 has been researched as a drug target for anti-virus drugs and allergy drugs; however, how it recognizes DNA was previously unknown.
This group of researchers clarified the steric constitution of three forms of TLR9 and found that TLR9 and microoranism-derived DNA sequences bind at a rate of 2 to 2 and form an activated dimer and DNA sequence is recognized by binding to the groove at the end of TLR9. DNA binding to TLR9 clarified by this group may develop vaccine adjuvant and therapeutic drugs for virus infection and allergy.
Abstract
Innate immunity serves as the first line of defense against invading pathogens such as bacteria and viruses1. Toll-like receptors (TLRs) are examples of innate immune receptors, which sense specific molecular patterns from pathogens and activate immune responses2. TLR9 recognizes bacterial and viral DNA containing the Cytosine-phosphate-Guanine (CpG) dideoxynucleotide motif3,4. The molecular basis by which CpG-DNA elicits immunostimulatory activity via TLR9 remains to be elucidated.
Here we show the crystal structures of three forms of TLR9: unliganded, bound to agonistic CpG-DNA, and bound to inhibitory DNA (iDNA). Agonistic CpG-DNA–bound TLR9 formed a symmetric TLR9/CpG-DNA complex with 2:2 stoichiometry, whereas iDNA-bound TLR9 was a monomer. CpG-DNA was recognized by both protomers in the dimer, in particular by the N-terminal fragment (LRRNT–LRR10) from one protomer and the C-terminal fragment (LRR20–LRR22) from the other. The iDNA, which formed a stem-loop structure suitable for binding by intramolecular base pairing, bound to the concave surface from LRR2–LRR10. This structure serves as an important basis for improving our understanding of the functional mechanisms of TLR9.
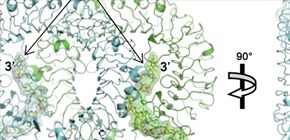
Fig. 1 Binding modes of TLR9.
(Upper) Dimer structure of the EcTLR9/DNA_CpG motif complex.
(Lower) Structure of EcTLR9 in complex with antagonist DNA.
In the dimer structure, EcTLR9 and its dimerization partner EcTLR9 are shown in green and cyan, respectively. The CpG motif has extended structure and binds to TLR9 dimer at two positions (2:2 complex), while antagonist DNA takes loop structure and binds to the interior of horseshoe shaped TLR9 molecule (1:1 complex).
To learn more about this research, please view the full research report entitled " Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9 " at this page of the Nature website.
Related Link
