Unraveling mystery of evolution of vertebrates by genome sequencing of goldfish
Toward elucidation of genome duplication and evolution of genes using next-generation DNA sequencing
A group of researchers from the Osaka University Institute for Protein Research, in cooperation with the National Institute of Genetics, Aichi Fisheries Research Institute, and National Institutes of Health (US), sequenced the goldfish genome, a world first.
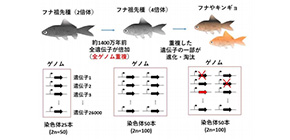
The goldfish ( Carassius auratus ), a cyprinid teleost closely related to the crucian carp, has had dozens of breeds developed since its domestication in China some 1,000 years ago.
Because of its diversity in shape, the goldfish is thought to be an excellent genetic model system for understanding the evolution of body shape in vertebrates, including humans.
To clarify the whole picture of genes related to diversity and body morphology, it is necessary to obtain complete genome sequences; however, the goldfish’s genome structure was too complicated to decode complete genome sequences. This is because the goldfish shares the same whole-genome duplication (WGD) event (allotetraploid event); that is, it is a hybrid of two closely related species created with both chromosome sets of each parent present in its gametes.
In order to overcome this challenge, the researchers used 71X PacBio long reads. This sequencing technology offers read length of more than 100 times that of conventional sequencing technology, allowing one to read 10,000~40,000 bases at a time, making it easy to create a genetic map based on more correct genome information. In addition, using the PacBio long reads, they generated a high-quality draft sequence of a Wakin goldfish, which has a body shape similar to that of wild crucian carp.
Also, by conducting a special treatment in the developmental stage, they produced goldfish in which all the genetic information originates from the female parent for use in their study. Taking this inventive approach, they simplified the genome structure, which in turn simplified DNA sequencing.
Their studies revealed:
- 1) The WGD event occurred in the goldfish ancestor genome 14 million years ago,
- 2) after the WGD, 12% of the genes in the goldfish genome were quickly lost, and
- 3) the expression of 30% of the retained duplicated gene diverged across tissues.
Genes easily affected by genetic selection and gene expression changes were also clarified. Genes were lost at a rate of 0.43% per million years (Ma) in goldfish, compared to 0.25% per Ma in salmon during the 80 Ma following the WGD. This means that goldfish are in an evolutionary process in which genes are lost 1.7 times faster than salmon during the 80 Ma following the WGD.
Genome sequencing by this group has opened the door for advancing research on body morphology, genome duplication, and evolution. Since some breeds of goldfish exhibit disease symptoms similar to those of human diseases, it is anticipated that goldfish will help to elucidate causes of diseases and establish diagnostics and therapeutics of human diseases. Thus, genome research has drawn attention from overseas, including the U.S.
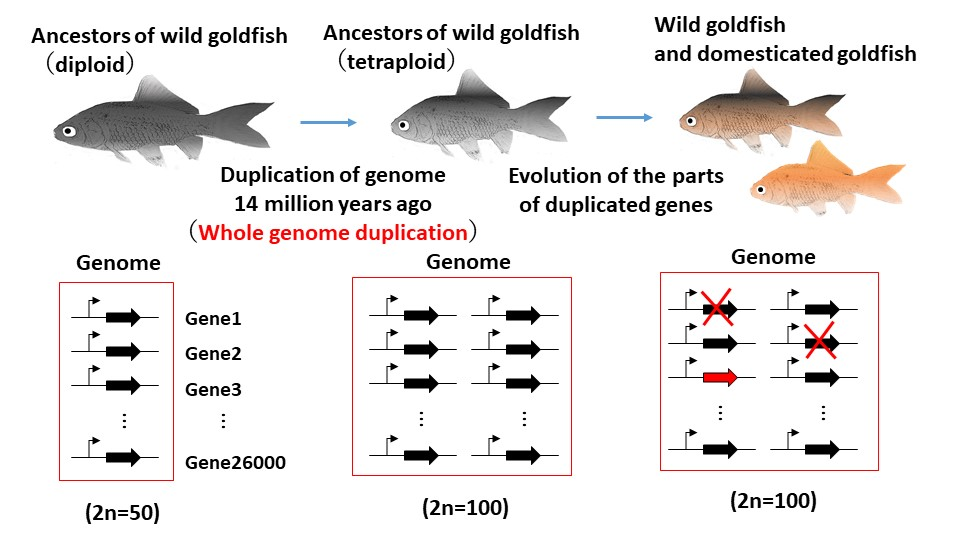
Figure 1 . The whole genome duplication (WGD) event occurred in the genome of goldfish ancestors 14 million years ago. The duplicated genes evolved after WGD. The expression of 30% of the retained duplicated gene diverged across tissues. 12% of duplicated gene pairs have lost one copy after WGD.
The article, "De Novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes afterwhole genome duplication" was published in Science Advances at DOI: https://doi.org/ 10.1126/sciadv.aav0547 .
Related links
