Switching off the light to see better
Researchers at Osaka University employ fluorescent molecules that can switch on and off, along with selective illumination in a plane, to create sharper microscope images that can be used to visualize internal cell structures or cell clusters
To study living organisms at ever smaller length scales, scientists must devise new techniques to overcome the so-called diffraction limit. This is the intrinsic limitation on a microscope’s ability to focus on objects smaller than the wavelength of light being used. Structured illumination microscopy is one of the super-resolution techniques that can help, by illuminating sinusoidal-patterned light on a sample to obtain a sharper image. However, background light from out-of-focus regions can still smear out the final picture.
In a study recently published in the journal Nature Methods, researchers from Osaka University demonstrated a new approach for super-resolution microscopy capable of observing structures inside a single cell or a cell cluster. This was accomplished by selecting only a desired plane to image using thin “light sheet” illumination, projected perpendicular to the lens, to switch on fluorophores. “We show that selective-plane activation allows us to image dense microstructures inside cells with excellent sharpness not readily available previously,” lead author of the study, Kenta Temma says. That is, sinusoidal “structured” light selectively excited only a thin plane where on-state fluorophores were localized, which allowed for background-free super-resolution imaging.
While some previous methods utilized random fluorescence emission from single molecules, or “donut” shaped second light source to deactivate or deplete fluorescent sources outside of a desired area, this new method can be gentler on cells that might be damaged by intense or long exposure to light. The researchers believe that their approach is especially effective when trying to understand what is happening in living systems with spatial structure, which can often exhibit background light outside the desired focal plane. This includes organoids, which are artificial assemblies of different cell types meant to reproduce the behavior of actual body organs much better compared with collections of cells cultured on a flat petri dish. “We anticipate that our technique will be useful for future biological studies of 3D cell clusters, including organoids,” says senior author, Katsumasa Fujita. The same could apply to other complex biological systems.
Fig. 1
Configuration and imaging schemes of SPA-SIM (selective plane activation structured illumination microscopy). Simulated distribution of effective excitation pattern in SPA-SIM
Credit: K. Temma, R. Oketani et al
Fig. 2
3D projection images of a living HeLa cell expressing Skyla-NS on mitochondria observed with SPA-SIM(proposed method) and conventional 3DSIM.
Credit: K.Temma, R.Oketani et al
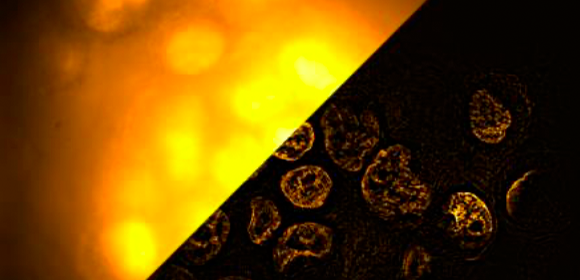
Fig. 3
Fluorescence images of nuclei in a cell spheroid labeled by rsGamillus-S with a diameter of 100 µm observed with widefield, SPA-SIM, and 3DSIM at a depth of 43 µm.
Credit: K.Temma, R.Oketani et al
The article, “Selective plane activation structured illumination microscopy,” was published in Nature Methods at DOI: https://doi.org/10.1038/s41592-024-02236-3.
Related Links
- Fujita Laboratory, Graduate School of Engineering, Osaka University
- Nagai Laboratory, SANKEN (The Institute of Scientific and Industrial Research), Osaka University
- Institute for Open and Transdisciplinary Research Initiatives, Osaka University
- Center for Future Innovation, Graduate School of Engineering, Osaka University

