
Study reveals genetic basis of quantitative traits and diseases in Japanese
Genome-wide association study reveals genetic basis of quantitative traits and complex biological links
Genome-wide association studies (GWAS) are an emerging method for scientists to identify genes involved in human disease. GWAS searches the whole genome region for small variations, called single nucleotide polymorphisms (SNPs), which occur more frequently in people with a particular disease than in people without. Each study can look at millions of SNPs at the same time. Researchers use data from this type of study to pinpoint genes that may contribute to a person’s risk of developing a certain disease.
Most existing GWAS have primarily examined European-ancestral subjects, and each separately focused on a few quantitative traits. To obtain a comprehensive landscape, additional studies in non-European populations are needed – and a team of Japanese researchers centered at Osaka University recently did just that.
“We conducted a large-scale GWAS using genetic information of more than 160,000 Japanese from the BioBank Japan Project,” explains Masahiro Kanai, lead author of the study recently published in Nature Genetics . “We successfully identified 1,407 genetic variations in the genome sequence that affect 58 traits, including anthropometric, metabolic, kidney-related, hematological, and blood pressure.”
The BioBank Japan Project, launched in 2003, enrolled 200,000 patients with 47 target diseases, making it one of the largest hospital based biobanks in the world. Without prior biological knowledge of cross-phenotype relationships, the team’s findings demonstrated that genetics clinical measurements successfully recapture their relevance to diseases, and thus could contribute to elucidation of unknown etiology and pathogenesis.
“By incorporating the additional GWAS results of the 32 complex diseases and traits in Japanese, we further identified cellular tissues that affect diseases and clinical laboratory values by cross-cutting omics analysis (that integrates with epigenome information) obtained from 220 types of cellular tissues,” co-senior author Yukinori Okada says.
The new findings also suggested there are complex interrelations between the clinical measurements and diseases, demonstrating the value of conducting GWAS for a variety of traits in a single large-scale cohort with detailed clinical information.
“Our study substantially expanded the knowledge of genetic relationships across clinical measurements and diseases,” co-senior author Yoichiro Kamatani adds. “Our results not only shed light on numerous clinically meaningful genetic mutations, but also showed that the integration of genomic information and epigenome information makes it possible to clarify the molecular and cellular basis of diseased tissues.”
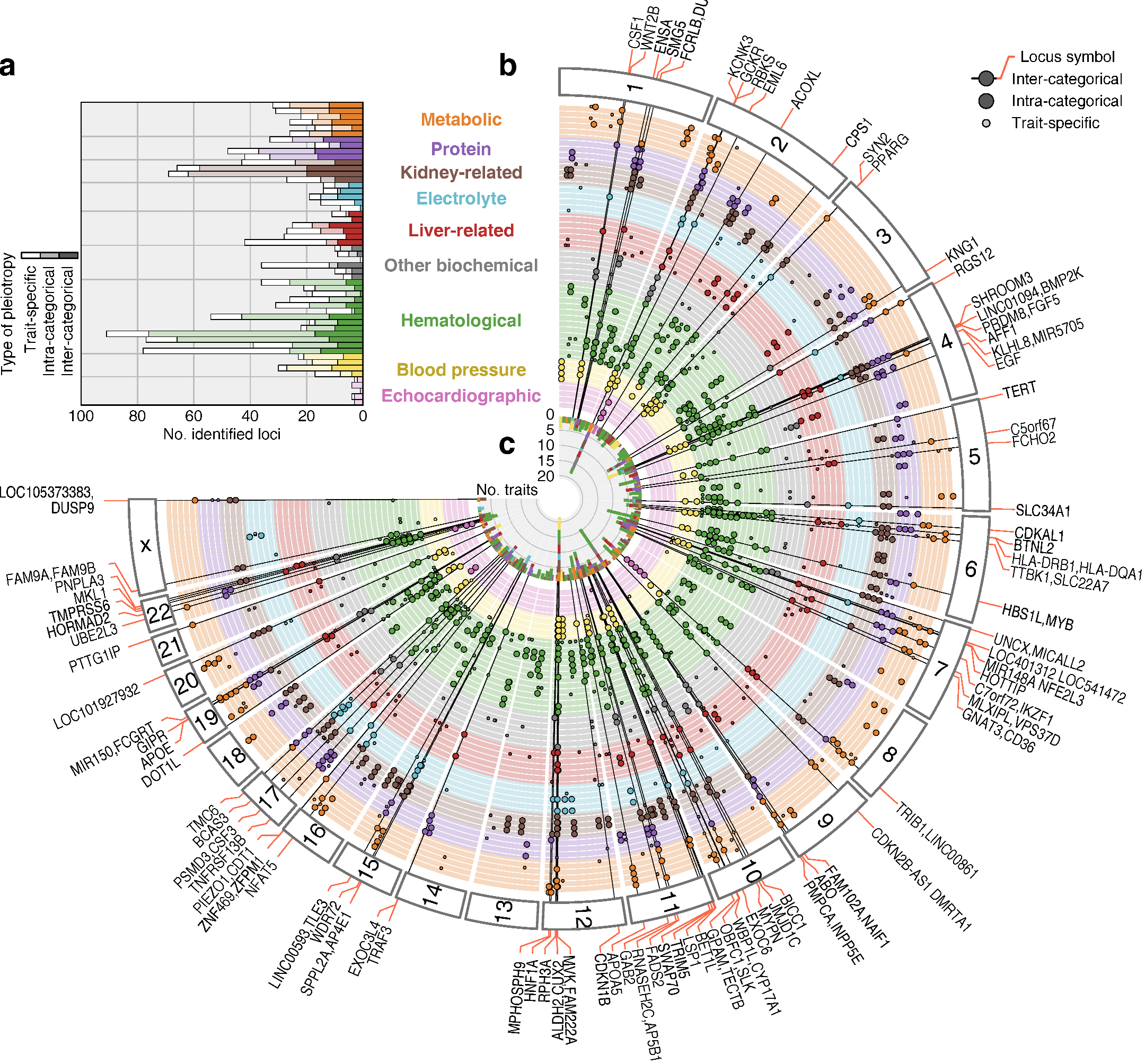
Fig.1 GWAS results of 58 quantitative traits for more than 160,000 Japanese subjects

Fig.2 Cell specificity of the quantitative traits identified by trans-omics analysis integrating the GWAS results and epigenetic data

Fig.3 Network among quantitative traits, human diseases, and cell types.
To learn more about this research, please view the full research report entitled " Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases " at this page of Nature Genetics .
Related links
