Peering into the genome of brain tumor
Osaka University-led research introduces a machine-learning algorithm for predicting genetic mutations of malignant brain tumor types based only on MRI, offering promise for noninvasive diagnosis and faster treatment
Researchers at Osaka University have developed a computer method that uses magnetic resonance imaging (MRI) and machine learning to rapidly forecast genetic mutations in glioma tumors, which occur in the brain or spine. The work may help glioma patients to receive more suitable treatment faster, giving better outcomes. The research was recently published in Scientific Reports .
Cancer treatment has undergone a revolution in recent years. Spurred by recognition that each cancer case is unique, the specific genetic mutations tumor cells carry are now sequenced to discover which chemotherapy drugs will work best. However, certain types of cancer, especially brain tumors, are less accessible for genetic testing. The tumor’s genotype can’t be found until a sample is taken during surgery, and this can significantly delay treatment.
Glioma is a type of cancer that originates in the brain’s supporting cells. Two types of mutations are especially important; these are changes in the gene for the enzyme isocitrate dehydrogenase (IDH) or the promoter region of telomerase (TERT). Identifying these mutations can help direct the proper course of treatment. The researchers produced a machine-learning algorithm that can predict which mutations are present using only the MR images of the tumors.
“Machine learning is increasingly used to diagnose medical images. But our work is one of the first to even attempt to classify something as hidden as the genotype based on image data alone,” study first author Ryohei Fukuma explains. The algorithm was found to be significantly better at predicting the mutations compared with conventionally used radiomic features of the MR images, such as size, shape, and intensity.
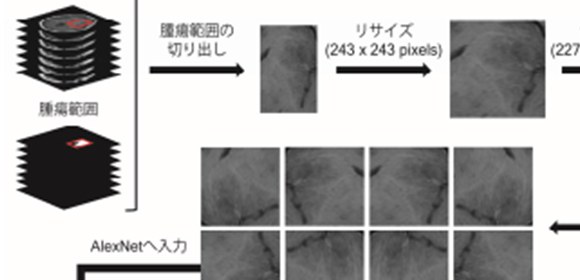
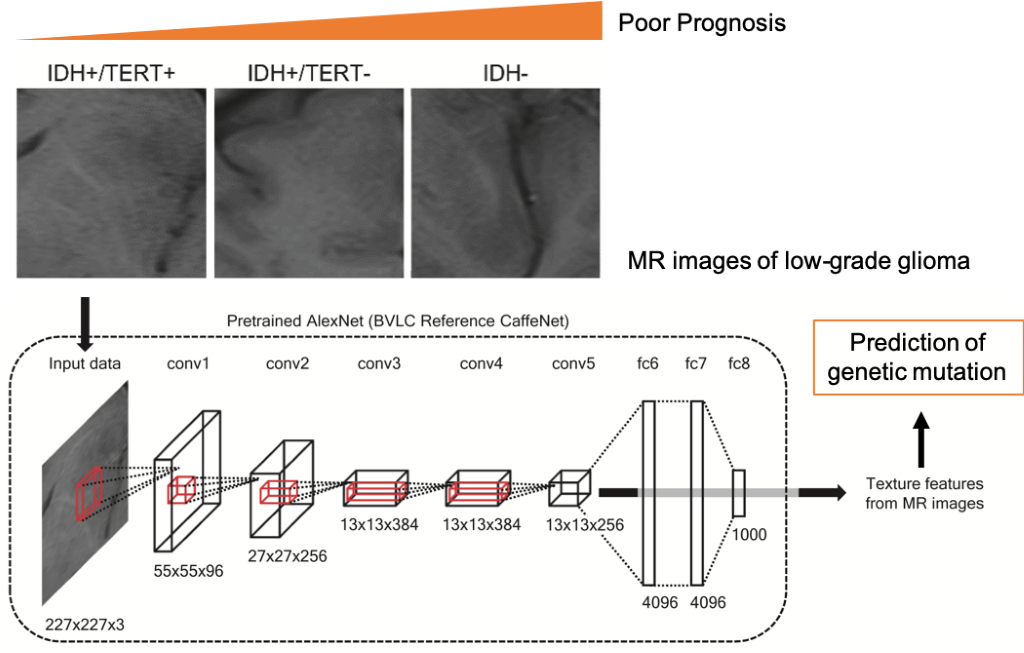
To construct the algorithm, the researchers used a convolutional neural network to extract features from the MR images. Then, using a machine-learning method called support vector machines, they classified the patients into groups based on the presence or absence of mutations. “We hope to expand this approach to other types of cancer, so we can take advantage of the large cancer gene databases already collected” senior author Haruhiko Kishima says.
The end result could remove the need for surgical tissue sampling. Even more, it could lead to better clinical outcomes for patients as the process of delivering personalized medicine becomes easier and faster.
Figure
MR images of gliomas were fed into a pre-trained deep neural network that can classify natural images. The activities of neurons in each layer of the network were used to characterize the textures of the MR images. These texture features were used to infer three key molecular subtypes of gliomas. The obtained algorithm was able to predict these molecular subtypes with an accuracy of 63.1%.
The article, “Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network” was published in Scientific Reports at DOI: https://doi.org/10.1038/s41598-019-56767-3 .
Related links
