Sleeping Beauty Helps Identify Genes Involved in a Fatty Liver-associated liver cancer
Researchers from Osaka University have used a gene screening technique called Sleeping Beauty mutagenesis to identify genes potentially responsible for a fatty liver-associated liver cancer
With an estimated twenty-thousand protein-coding genes in the human genome, pinpointing a specific gene or pathway responsible for a particular disease can be like finding a needle in the proverbial haystack. This has certainly been the case for hepatocellular carcinoma (HCC), with studies identifying more than ten thousand mutations in the cancer genomes of HCC patients, making it extremely difficult to develop targeted therapies.
In addition to being the second most lethal form of cancer worldwide, increasing numbers of people are diagnosed with HCC each year. A major emerging risk factor for this type of liver cancer, particularly in Western countries, is non-alcoholic fatty liver disease (NAFLD), characterized by excess fat storage in the liver. Linked to a high-fat diet and type 2 diabetes, damage caused to the liver by NAFLD is likened to that seen in cases of serious alcohol abuse.
Given the lack of treatment options and poor prognosis for HCC patients, research teams led by Osaka University set out to identify genes driving the development of NAFLD-HCC. “We used a new technology to comprehensively search for oncogenes in individual animals by large-scale screening of genes and signal transduction pathways that contribute to the development of liver cancer associated with fatty liver disease” explains Tetsuo Takehara, co-author of a recent study published in Proceedings of the National Academy of Sciences of the United States of America .
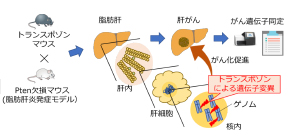
This new technology involves small, mobile DNA sequences—charmingly named Sleeping Beauty transposons—that were randomly integrated into the genome in two separate mouse models of NAFLD. Transposon insertion disrupts genes normally involved in tumor suppression or activates nearby oncogenes, allowing researchers to identify potential HCC-causing genes by accelerated or excessive liver tumor development in affected mice.
“Amongst hundreds of candidate genes, we discovered that Sav1, a component of the Hippo pathway, was the most frequently mutated gene in both Sleeping Beauty screens” says leading author Takahiro Kodama. Interestingly, Sav1 and the Hippo pathway are involved in the regulation of organ size. If this pathway is disrupted, liver progenitor cells replicate uncontrollably, leading to tumor formation.
The researchers noted significant liver damage and accumulation of proteins involved in tumor formation in NAFLD mice with a deletion of the Sav1 gene, suggesting they had found their “needle”. “Although we knew that deletion of Sav1 leads to liver enlargement and the development of liver tumors, these new findings provide a key link between dysregulation of the Hippo pathway and the development of HCC in NAFLD” says Kodama. “Knowing this, we can potentially develop new liver cancer treatments targeting the Hippo pathway.”
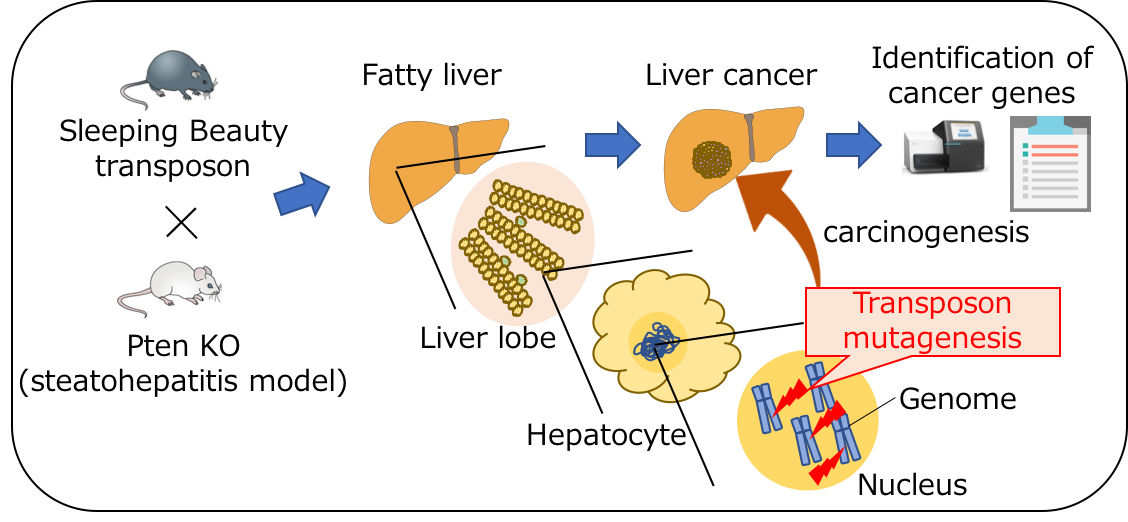
Fig.1: Genome-wide cancer gene discovery by Sleeping Beauty transposon mutagenesis
(credit: Osaka University)

Fig.2: Sav1 gene plays an important protective role in development and progression of fatty liver-associated hepatocellular carcinoma.
(credit: Osaka University)
The article, “Molecular profiling of non-alcoholic fatty liver disease-associated hepatocellular carcinoma using SB transposon mutagenesis”, was published in Proceedings of the National Academy of Sciences of the United States of America at DOI: https://doi.org/10.1073/pnas.1808968115 .
Related links
