Crystal structures of M. pneumoniae proteins play a critical role in infection
A group of researchers from 9 research institutes in Japan, Barcelona (Spain), and Frankfurt (Germany), including Osaka Metropolitan University, Osaka University, and the National Institute of Infectious Diseases, has clarified the atomic structure of a transmembrane complex Nap of Mycoplasma pneumoniae (M. pneumoniae).
M. pneumoniae, a bacterial human pathogen that causes primary atypical pneumonia, binds to sialylated oligosaccharides on the human epithelial surfaces, gliding on host cell surfaces, and initiating the infectious process. Every year, tens to hundreds of thousands of people develop M. pneumoniae pneumonia in Japan. M. pneumoniae pneumonia accounts for 10 to 30% of human community-acquired pneumonia, and an outbreak of M. pneumoniae pneumonia occurred in 2010~2011.
M. pneumoniae forms a polar structure known as the attachment organelle and glides in the direction of this differentiated structure. M. pneumoniae tightly binds to host target cells through its attachment organelle. The Nap on the surface of the attachment organelle is composed by the immunodominant proteins P1 and P40/P90, which mediate M. pneumoniae motility and infectivity.
M. pneumoniae cells bind to human epithelial surfaces through sialylated oligosaccharides. For M. pneumoniae, avoiding the human immune system by adhesion, gliding, and conformational changes is necessary for maintaining its infectivity and survival. Nap, which plays an important role in adhesion, gliding, and conformational changes of M. pneumoniae had drawn attention since its discovery around 1980, but its structure was unknown.
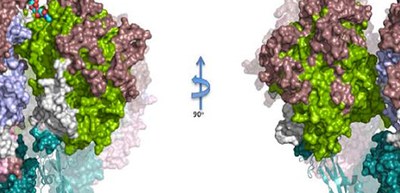
In this study, the researchers determined the atomic structure of the Nap protein P1 extracellular region, clarifying the whole structure of P1 and P40/P90 by X-ray crystallography and cryo-electron microscopy. The structure looked like a bundle of four corn dogs on a stick stuck into the mycoplasma membrane. The binding site for sialic acid was found in P40/P90 (in a pocket at the top of the Nap) and not in P1.
Regions with genetic variability of P1 and P40/P90 cover most of the surface from the upper part of the Nap and genetic and clinical variability concentrates on the N-terminal domain (top) surfaces of P1 and P40/P90.
The structure of M. pneumoniae clarified by this group is important information for understanding the functions of Naps and the mechanisms behind adhesion, gliding motility, and conformational changes of M. pneumoniae as a strategy to evade the immune host system.
This group’s achievements will significantly promote atomic-level understanding of adhesion and gliding motility of M. pneumoniae. This will allow scientists to design chemical compounds that inhibit adhesion of M. pneumoniae, and identification of a relatively weak or poorly accessible antigenic region will open up new possibilities in vaccine development against M. pneumoniae infections.
Reference
The article, “Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae,” was published in Nature Communications at DOI: https://www.nature.com/articles/s41467-020-18777-y.
