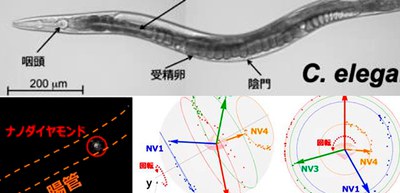
3-DoF rotation tracking method using nanoscale sensor
A group of researchers from the National Institutes for Quantum Science and Technology (QST), Hiroshima University, Osaka University, and Kyoto University has demonstrated that a three-degrees-of-freedom (3-DoF) rotation of biomolecules can be visualized via nitrogen-vacancy centers (NV centers) in a fluorescent nanodiamond.
A NV center, a point defect in diamond crystal lattice, consists of a substitutional nitrogen atom and a neighboring vacancy. Since its electronic spin is highly sensitive to an external magnetic field under ambient conditions, the NV center has been used for sensing and has drawn attention as a promising quantum sensor for physics and biology.
The rotation of an object cannot be fully tracked without understanding a set of three angles: roll, pitch, and yaw. Tracking these angles as a 3-DoF rotation is a fundamental measurement. High resolution fluorescent microscopes can pick up the translational motion of biomolecules; however, there has been no method to track 3-DoF rotation to measure nanometer-scale dynamics in biomolecules and live cells.
The researchers of this group established a method to measure 3-DoF rotation, in conjunction with an Optically Detected Magnetic Resonance (ODMR) spectrometric microscope, using the spin of nanodiamonds as an orientation sensor. They demonstrated that 3-DoF rotation of biomolecules could be visualized via nitrogen-vacancy centers in a fluorescent nanodiamond.
The spin state of an NV center correlates with electromagnetic fields and can be readout by ODMR by measuring spin-state-dependent fluorescence intensity. Thus, this group developed a method to determine the angle of spins in the NV center in a nanodiamond by externally applying a magnetic field with arbitrary magnitude and direction to determine the orientation of a nanodiamond in the space of a rigid body.
They also showed that, in conjunction with an ODMR spectrometric microscope, the spin of nanodiamonds can be used as an orientation sensor to measure nanometer-scale 3-DoF dynamics in vitro, in cells, and in vivo. They successfully applied our 3-DoF rotation tracking method to the visualization of single-molecule conformational changes, cellular membrane dynamics, and nanometer-scale motion in live animals.
Specifically,
1. They tracked the rotation of a single molecule of the motor protein F1-ATPase, an enzyme called ATPase, by attaching a nanodiamond to the γ-subunit, and found that the γ-subunit shaft rotated 120° after each step of ATP hydrolysis and showed a 3-step rotation for each revolution cycle.
2. They probed the dynamics of the plasma membrane of cells of the epidermoid carcinoma model cell line A431, which expresses a high concentration of epidermal growth factor receptors (EGFRs), showing that 3D rotational motion of the membrane protein correlated with intracellular cytoskeletal density.
3. They applied 3-DoF rotation tracking to the visualization of the intestine of live nematode Caenorhabditis elegans (C. elegans).
This group’s 3-DoF rotation tracking method for visualizing the movements of molecules in living cells in real time will clarify mysteries in life. Since the binding of drugs to a protein target changes the structure and rotation of the protein, the development of drugs that can change the rotational degrees of freedom of interacting partners will be a new strategy for drug discovery.
Figure 1
Figure 2
Figure 3
Figure 4
Figure 5
The article, “Tracking the 3D Rotational Dynamics in Nanoscopic Biological Systems” was published in Journal of the American Chemical Society at DOI: https://pubs.acs.org/doi/10.1021/jacs.0c01191.
