Hepatitis C virus (HCV) sneaks into the active protein translation of a host cell
- Hepatitis C virus (HCV) is the cause of hepatitis C and some cancers such as liver cancer.
- The precise molecular mechanism by which HCV induces protein production in a host cell remains elusive.
- Researchers identified a new mechanism of HCV-induced protein production by using cutting edge technology such as cryo-electron microscopy (Cryo-EM) and single molecule imaging.
- The findings could contribute to the development of a new antiviral treatment.
2. Summary
Researchers at RIKEN, University of Hyogo and Osaka University identified a novel mechanism that allowed the Hepatitis C virus (HCV) to produce its own protein efficiently, in the host cell.
This finding could contribute to the development of a new antiviral treatment.
HCV is the cause of hepatitis C and some cancers such as liver cancer. The RNA genome of HCV induces viral protein production in the host cell. During protein production procedure (translation), the RNA genome has to be decoded into protein by the host's ribosomes (protein production machinery). From the past studies, the special region of RNA genome called IRES, Internal Ribosome Entry Site, has been known to be important for translation of HCV RNA genome. However, how IRES contributes to enhance the protein production remains elusive.
Here, researchers used cutting edge techniques such as the cryoelectron microscopy (Cryo-EM) and single molecule imaging to visualize how the HCV RNA genome sneaks into the host protein production system. They identified that the IRES part of the HCV RNA genome binds to the active ribosome without hindering the on-going translation. Furthermore, after the on-going translation terminates, the HCV RNA genome hijacks the ribosome and forces it to produce the viral protein in the next round of translation. This hijacking mechanism seems to be conserved in similar RNA viruses, suggesting a source of the powerful infection ability of the RNA virus.
This work was published on-line in the American scientific journal “Molecular Cell” on May 9th, 2019.
3. Background
A virus has its own genome (made of either DNA or RNA) and capsid for the genome storage. However, a virus cannot self-propagate and requires the host cell for replication. Thus, it has many sophisticated mechanisms to enhance its replication process. With these replication process, some viruses are known to damage the life cycle of the host cell. HCV is one example of such viruses. It causes hepatitis C and some cancers such as liver cancer. HCV has single strand RNA as its genome.
Genome information of certain virus is decoded using the sequence information of RNA, and the process of using RNA information to connect amino acids and synthesized protein is called "Translation". Translation reaction is a very complicated process and thus half of the total energy of a cell is consumed by translation. During its lifecycle, a virus has to make many viral capsids to propagate. Thus the virus has to force the host cell's translation machinery called “ribosome” to produce viral proteins. The eukaryotic ribosome is a huge protein-RNA complex, and has two subunits called “large (60S)” and “small (40S)” subunits.
As a marker for translation, eukaryote host RNA has a "cap structure", where tens of translation initiation factors and the 40S and 60S ribosomal subunits form an initiation complex, and the translation process begins from the translation initiation site. However, some kinds of viruses such as HCV do not have the "cap structure", but use IRES and a smaller number of initiation factors to start the translation process. IRES is a special region located at the end of the HCV RNA genome. IRES contribute to the efficient translation of the HCV RNA genome; however, the precise mechanism by which IRES does this remains elusive. This is because, the past study lack the information of IRES during the active translation.
4. Results
IRES was thought to bind to the freely diffusing small (40S) ribosomal subunit and then participate in the translation process based on the structural insights from previous studies. However, whether IRES can bind to the actively translating ribosomes, which contain the on-going host RNA, remained elusive. Surprisingly, our research found that the IRES can bind to the actively translating ribosomes of a human host’s cell.
To reveal the mechanism of IRES mediated ribosomal translation, researchers reconstituted the eukaryotic translation system in a test tube (in vitro assay). Using the cutting edge Cryo-EM, they found that IRES can bind to the specific sites of active ribosomes without hindering the on-going transcription (Figure 1).
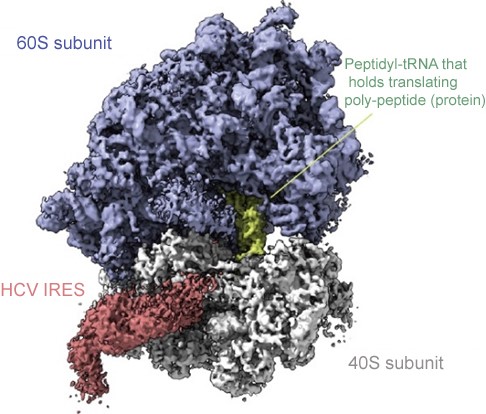
Figure 1 | Cryo-EM image of an active ribosome with bound HCV RNA genome.
The HCV genome RNA binds to a specific site that does not hinder the on-going translation process. IRES, Internal Ribosome Entry Site, part of the HCV RNA genome is colored red, and peptidyl-tRNA that holds translating poly-peptide (protein) is colored green.
Moreover, single molecule imaging confirmed that IRES can actually bind to the actively translating ribosome. These results indicate that IRES sneaks onto an actively translating host ribosome, waits for the termination of the on-going host cell's translation, and then hijacks the ribosome to translate the viral protein.
Biochemical analysis revealed that the efficiency of viral protein production is much higher when the HCV RNA genome is mixed with actively translating ribosomes than when mixed with vacant freely diffusing ribosomes. Thus, taking all the data into consideration, our data suggests that the new "hijack" model (scheme I→II→III→IV→V in Figure 2, HCV RNA binds to an on-going active ribosome) proposed here is more efficient for viral protein production than the traditional model (scheme VI→III→IV→V in Figure 2, HCV RNA binds to a vacant ribosome). Furthermore, the new pathway also suggests a possibility of reuse of the small (40S) subunit (scheme V→III).
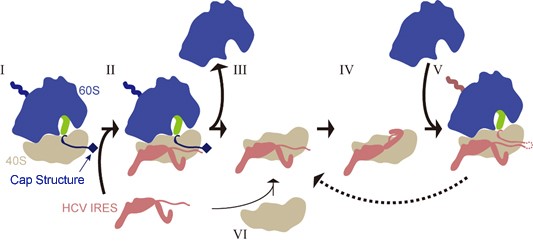
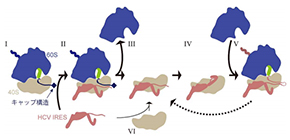
Figure 2 | Hijacking scheme of the HCV RNA genome.
Schematic illustration of the translation process by 40S (small subunit, gray), 60S (large subunit, blue), peptidyl-tRNA (green), mRNA with cap structure (black) and the HCV RNA genome (brown). First the HCV RNA genome binds to an actively translating ribosome (scheme I→II), and then, after termination of translation, 40S and 60S ribosomal subunits are separated (scheme III), and the HCV RNA genome forces the ribosome to translate HCV RNA and produce viral protein (scheme IV→V). In certain cases, the same 40S-HCV RNA complex is maintained and works together in the next round of translation (scheme V→III) as well. HCV RNA genome may also bind to a freely diffusing 40S subunit instead of binding to an active ribosome (scheme VI).
5. Conclusion
One of the advantage of Japanese structural biology is the unique combination of cutting edge technology (e.g. Cryo-EM and Single molecule imaging) with classical biochemistry techniques such as in vitro reconstitution. With this advantage, we succeeded in finding a new "hijacking" mode of the HCV RNA genome. This mechanism would be common to various virus. Therefore, if we can develop an inhibitor against the HCV RNA - ribosome interaction, it might be an effective anti-viral therapeutic.
Translation is one of the most fundamental reactions in a cell. Here, we used a unique combination of cutting edge imaging technologies and traditional sample preparation and assay technologies. Although "translation" is a classical theme, like this hijacking mechanism, there might be other hidden mechanisms that may have been overlooked in the past. We hope that our multidisciplinary approach will contribute to unveil the mysteries of the biology.
6. Acknowledgement
This research was supported by Grants-in-Aid for Scientific Research on Innovative Areas "Nascent Chain Biology" (JP15H01548 and JP17H05677 to T.I. and JP26116002 to H.I.), "Live-Protein Dynamics" (JP15H01656 and JP17H05897 to H.S.) and "Noncoding RNA Neo-taxonomy" (JP26113007 to H.T.) and Grants-in-Aid for Scientific Research (B) (JP16H04756 to T.I., JP15H04324 to H.I., and JP16KT0068 to H.T.) from the Japan Society for the Promotion of Science (JSPS), Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) (JP18am0101082) from the Japan Agency for Medical Research and Development (AMED), the Takeda Science Foundation, the RIKEN Pioneering Project "Dynamic Structural Biology" and the Cooperative Research Program of the Institute for Protein Research, Osaka University (CRa-18-01 to H.T. and T.I.).
7. Publication information
<Title>
HCV IRES captures an actively translating 80S ribosome
<Authors>
Takeshi Yokoyama, Kodai Machida, Wakana Iwasaki, Tomoaki Shigeta, Madoka Nishimoto, Mari Takahashi, Ayako Sakamoto, Mayumi Yonemochi, Yoshie Harada, Hideki Shigematsu, Mikako Shirouzu, Hisashi Tadakuma*, Hiroaki Imataka*, and Takuhiro Ito* (*Corresponding authors)
<Journal>
Molecular Cell
8. Key words
[1] Hepatitis C virus (HCV)
HCV genome is single strand RNA and codes three capsid structure proteins (Core, E1 and E2) and seven non-capsid proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B). Normally, HCV only infect to humans and chimpanzees. In the world, approximately 170 million people are carriers of the HCV.
[2] Ribosome
Decodes genetic information stored in the RNA to the polymer of the amino acid sequence, the protein. Thus, the ribosome serves as a protein production factory of the cell. The core of a ribosome is composed of RNA and the RNA core is covered by ribosomal proteins. Small and large subunits of the eukaryotic ribosome are called 40S and 60S subunits.
[3] IRES (Internal Ribosome Entry Site)
In the normal eukaryotic translation process, initiation factors recognize the 5' cap structure and recruit ribosomes. On the other hand, in the viral protein production, IRES skips the complicated cap-dependent initiation process and thus, the initiation process of the viral protein has been thought to be more efficient than the host protein production. However, in the past, there has not been direct confirmation of this process and the details had remained elusive.
[4] Cryoelectron microscopy (Cryo-EM)
Quick freezing by liquid ethane allows us to confine biomolecules within the thin layer of glass state water, where biomolecules maintain their intact structure. Researchers can reconstitute the high resolution three dimensional (3D) structures of biomolecules by observing the cryo-samples with a transmission electron microscope and by processing of the captured images with the help of an image processing technology.
[5] Single molecule imaging
Using florescent microscopy that can detect a single molecule of a fluorophore, researchers can observe the dynamics of biomolecules in real time.
[6] Cap structure of RNA
Most of the RNA that can be translated by the ribosome has a guanosine-modified 5' cap structure. In addition to the contribution of the cap structure to translation initiation, it also increases the stability of the RNA.
[7] Initiation factors (IFs)
IFs recognize the cap structure and recruit ribosomes. Eukaryotic IFs are called eIF (eukaryotic Initiation Factor).
9. URL of presenters
RIKEN, Ito Laboratory
http://www.riken.jp/en/research/labs/bdr/transl_struct_biol/
Hyogo University, Imataka Laboratory
http://www.eng.u-hyogo.ac.jp/group/group37/indexEnglish.html
Osaka University, Harada Laboratory
https://www.ccc.osaka-u.ac.jp/protein/nanobiology/
10. Contributed researchers of Osaka University
Yoshie Harada
Hisashi Tadakuma
Related links
- Harada Laboratory, Institute for Protein Research, Osaka University (link in Japanese)
