Winning the Arms Race: Analysis Reveals Key Gene for Bacterial Infection
Researchers at Osaka University use molecular-level analyses of evolution to identify a key gene in bacterial infection, potentially providing a novel target for drugs that could minimize the incidence of bacterial resistance
To successfully infect their hosts, bacteria need to evade the host immune system in order to reproduce and spread. Over the course of evolution, hosts—such as humans—develop increasingly sophisticated defenses against bacterial infection, while bacteria in turn develop new strategies to overcome these defenses in a biological arms race.
This competition has led to the development of antibiotic-resistant bacteria, for which few or no medical treatments are effective. Hence, there is an increasingly urgent need for novel drugs for use against such bacteria.
A research team at Osaka University have provided new hope in the battle against antibiotic-resistant bacteria by revealing a genetic factor that is important for the virulence of Streptococcus pneumoniae . This bacterium causes sepsis, pneumonia, and meningitis and is a major threat to public health globally. The team used molecular evolutionary analysis of gene sequences to identify a gene that has been largely prevented from mutating into other variant forms, suggesting that it is essential for the infection and/or reproduction of this bacterium.
In this work, reported in the journal Communications Biology, the team focused on genes encoding proteins called choline-binding proteins (CBPs), which are present on the bacterial cell surface and interact with the host’s immune system.
“We analyzed all the codons in genes encoding CBPs and compared their levels of diversity, reflecting how tolerant to mutation each gene is,” Shigetada Kawabata says. “We found that, in the gene cbpJ , over 13% of codons were under negative selection, meaning that if mutations had occurred in these regions, they reduced bacterial survival and led to the mutant being eliminated from the population.”
This finding led the team to investigate cbpJ ’s effects. They found that when mice were intranasally infected with wild-type or cbpJ -deficient S. pneumoniae , those with the mutant bacteria were more likely to survive and had fewer bacteria in their lungs.
“We then cultured the cbpJ -deficient S. pneumoniae with neutrophilsin medium and found that the mutant bacteria were generally less able to survive; however, when neutrophils were absent from the medium, they actually did better than the wild-type bacteria,” lead author Masaya Yamaguchi says. “This suggests that cbpJ helps bacteria to evade detection and clearance by neutrophils.”
The fact that the cbpJ gene is under strict negative selective pressure makes it a particularly attractive target for drugs, as this pressure would limit the likelihood of drug-resistant mutants emerging. This study also shows the value of molecular evolutionary analysis to identify new drug targets, including among pneumococcal virulence factors, especially when combined with traditional molecular microbiological approaches.
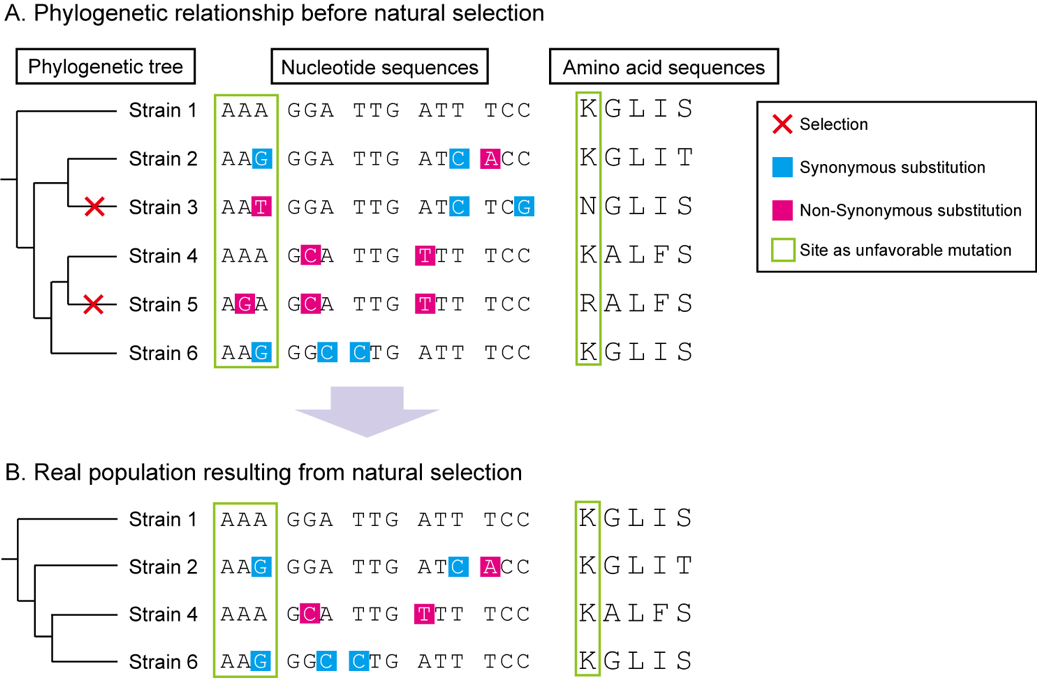
Figure: Scheme for intraspecies molecular evolutionary analysis
A. Random genetic drift induces synonymous and nonsynonymous mutations with equal probability. However, nonsynonymous mutations in essential regions are subjected to selective pressures. B. As a result of natural selection, synonymous substitutions are concentrated in important genes. Phylogenetic and molecular evolutionary analyses can detect significant accumulation of synonymous substitutions in codons of host proteins. Codon-based analysis yields much more information than nucleotide- or amino acid-based analyses.
(©️2019 Yamaguchi M. et al., Communications Biology)
The article “ Identification of evolutionarily conserved virulence factor by selective pressure analysis of Streptococcus pneumoniae ” is published in Communications Biology at doi: https://doi.org/10.1038/s42003-019-0340-7 .
Related links
