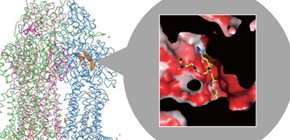
Clarification of structure of multidrug efflux proteins bound to inhibitors
a step toward overcoming Multidrug Resistant Pseudomonas aeruginosa (MDRP)
As part of Core Research for Evolutionary Science and Technology (CREST) of Japan Science and Technology Agency (JST), under the leadership of YAMAGUCHI Akihito , Specially Appointed Professor, Institute of Scientific and Industrial Research, Osaka University, a group of researchers succeeded in determining the structure of multidrug efflux proteins in Pseudomonas aeruginosa and E.coli bound to inhibitors.
Bacterial pathogens once thought to have been overcome by the development of antibiotics have become a big threat due to the emergence of multiple-drug-resistance. There are no therapeutic agents effective for Pseudomonas aeruginosa with multidrug resistance and the main cause for this lies in its multidrug efflux proteins that eject drugs as foreign substances. By clarifying the selective binding structure between multidrug efflux proteins and inhibitors, this group has paved a way toward overcoming Multidrug Resistant Pseudomonas aeruginosa (MDRP), a major medical threat.
Abstract
The multidrug efflux transporter AcrB and its homologues are important in the multidrug resistance of Gram-negative pathogens. However, despite efforts to develop efflux inhibitors, clinically useful inhibitors are not available at present. Pyridopyrimidine derivatives are AcrB- and MexB-specific inhibitors that do not inhibit MexY; MexB and MexY are principal multidrug exporters in Pseudomonas aeruginosa. We have previously determined the crystal structure of AcrB in the absence and presence of antibiotics. Drugs were shown to be exported by a functionally rotating mechanism through tandem proximal and distal multisite drug-binding pockets. Here we describe the first inhibitor-bound structures of AcrB and MexB, in which these proteins are bound by a pyridopyrimidine derivative. The pyridopyrimidine derivative binds tightly to a narrow pit composed of a phenylalanine cluster located in the distal pocket and sterically hinders the functional rotation. This pit is a hydrophobic trap that branches off from the substrate-translocation channel. Phe 178 is located at the edge of this trap in AcrB and MexB and contributes to the tight binding of the inhibitor molecule through a π–π interaction with the pyridopyrimidine ring. The voluminous side chain of Trp 177 located at the corresponding position in MexY prevents inhibitor binding. The structure of the hydrophobic trap described in this study will contribute to the development of universal inhibitors of MexB and MexY in P. aeruginosa.

To learn more about this research, please read the full research report entitled " Structural basis for the inhibition of bacterial multidrug exporters " at this page of the Nature website.
Related link :
