Designing antibody medicines using alanine substitutions
Will lead to the development of antibody drugs for treating cancer by enhancing antibody affinity by alanine substitutions that are used for analyzing functions of amino acid residues
Researchers from The University of Tokyo and Osaka University clarified a mechanism behind the antibody-antigen interface, developing a method for drastically improving antigen-binding affinity.
Designing antibodies that bind only to cancer cell-specific antigens to efficiently kill tumor cells is critical in developing antibody medicines for treating diseases such as cancer. Thus, the researchers tried to enhance the affinity of a cancer-targeted antibody, B5209B, to the fifth immunoglobulin domain (Ig5) of antigen ROBO1.
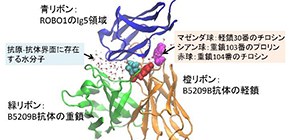
First, they obtained a complex structure consisting of the Ig5 domain of ROBO1 and the Fab of B5209B by X-ray crystallography, identifying which amino acid residue formed the antigen-antibody interface. Next, to study the importance of interfacial amino acid residues, they performed experiments to measure antigen-binding affinity, or Alanine scanning, by replacing the interfacial residues of a cancer-targeted antibody with Ala.
As a result, they identified important amino acid residues (hot spot residues) which cause deterioration of the high affinity by Ala substitution. They also discovered two amino acid residues (cold spot residues) which significantly enhanced antigen-binding affinity by Ala substitutions: the heavy chain P103 H and the light chain Y30 L of B5209B.
The P103 H A mutein, in particular, exhibited about 30 times higher affinity than the wild type (WT). The affinity enhancement was achieved by two types of substitutions: the binding affinity for the antigen was produced by a gain of binding enthalpy for heavy chain P103 H and by a gain of binding entropy for light chain Y30 L .
P103 H A substitution enhanced interaction energy between the antigen and the antibody. In fact, MD simulations showed that the interaction energy was larger for the P103 H A mutein than for the WT.
In addition, Y30 L A substitution enhanced the binding affinity for the antigen by reducing the number of water molecules trapped in the antigen-antibody interface. MD simulations showed that Y30 L A substitution enhanced the binding affinity for the antigen, which corresponded with enthalpic results obtained by thermodynamic analysis.
The results of this study will contribute to the development of antibody medicines for treating liver cancer and lung cancer through the analysis of B5209B. At the same time, the clarified molecular mechanism to strengthen antibody function will serve as guidelines for designing future antibodies, promoting rational design and development of antibody medicines.
Abstract
To investigate favorable single amino acid substitutions that improve antigen-antibody interactions, alanine (Ala) mutagenesis scanning of the interfacial residues of a cancer-targeted antibody, B5209B, was performed based on X-ray crystallography analysis. Two substitutions were shown to significantly enhance the binding affinity for the antigen, by up to 30-fold. One substitution improved the affinity by a gain of binding enthalpy, whereas the other substitution improved the affinity by a gain of binding entropy. Molecular dynamics simulations showed that the enthalpic improvement could be attributed to the stabilization of distant salt bridges located at the periphery of the antigen-antibody interface. The entropic improvement was due to the release of water molecules that were stably trapped in the antigen-antibody interface of the wild-type antibody. Importantly, these effects of the Ala substitutions were caused by subtle adjustments of the binding interface. These results will be helpful to design high-affinity antibodies with avoiding entropy-enthalpy compensation.
Figure 1. Schematic drawing of ROBO1 extra-cellular region and B5209B antibody
Figure 2. Crystal structure of B5209B antibody complexed with ROBO1 Ig5 antigen
The article, “Affinity Improvement of a Cancer-Targeted Antibody through AlanineInduced Adjustment of Antigen-Antibody Interface” was published in the Structure at DOI: https://doi.org/10.1016/j.str.2018.11.002.
Related links
