New methods of enhancing efficiency of genetic engineering in mice and rats developed
Able to modify genomes 100x larger than in previous methods
A group of researchers led by MASHIMO Tomoji (Associate Professor, Institute of Experimental Animal Sciences, Graduate School of Medicine) and YOSHITOMI Kazuto (Assistant Professor, Mouse Genomics Resource Laboratory, National Institute of Genetics, Research Organization of Information and Systems) developed two new gene modification methods: lsODN (long single-stranded oligodeoxynucleotide) and 2H2OP (two-hit two-oligo with plasmid). (Figure 1)
These methods use CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) -Cas systems and ssODN (single-stranded oligodeoxynucleotide).
CRISPR-Cas systems have made gene modification in mice and rats easy. By introducing Cas9 messenger RNA and gRNA, gRNA recognizes targeting DNA and Cas9 cuts the targeting site. DNA breaks are repaired by non-homologous end joining, which causes DNA mutations, resulting in gene knock-out.
Likewise, when ssODN is introduced with Cas9-gRNA, DNA breaks are repaired through homology-directed repair using donor DNA, resulting in knock-in of DNA sequences with one to dozens of bases (bp). However, ssODN allowed the synthesis of oligos up to 200 bp, therefore, which made it difficult to knock in large DNA sequences such as GFP (green fluorescent protein).
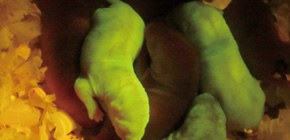
With these two gene modification methods, this group succeeded in achieving efficient and precise knock-in of GFP genes, the introduction of large genomic regions (approx. 200kbp), which was conventionally impossible, as well as replacing rat genes with human-derived genes, or generating gene humanized animals. (Figure 2)
These two knock-in methods will increase the efficiency of genetic engineering in mice and rats, as well as other various species of organisms, and will become very useful techniques for producing new genetically engineered organisms. It is highly anticipated that these genetically engineered organisms will be used in a wide field of studies such as drug development, translational research, and regenerative medicine.
Abstract
The CRISPR-Cas system is a powerful tool for generating genetically modified animals; however, targeted knock-in (KI) via homologous recombination remains difficult in zygotes. Here we show efficient gene KI in rats by combining CRISPR-Cas with single-stranded oligodeoxynucleotides (ssODNs). First, a 1-kb ssODN co-injected with guide RNA (gRNA) and Cas9 messenger RNA produce GFP-KI at the rat Thy1 locus. Then, two gRNAs with two 80-bp ssODNs direct efficient integration of a 5.5-kb CAG-GFP vector into the Rosa26 locus via ssODN-mediated end joining. This protocol also achieves KI of a 200-kb BAC containing the human SIRPA locus, concomitantly knocking out the rat Sirpa gene. Finally, three gRNAs and two ssODNs replace 58-kb of the rat Cyp2d cluster with a 6.2-kb human CYP2D6 gene. These ssODN-mediated KI protocols can be applied to any target site with any donor vector without the need to construct homology arms, thus simplifying genome engineering in living organisms.
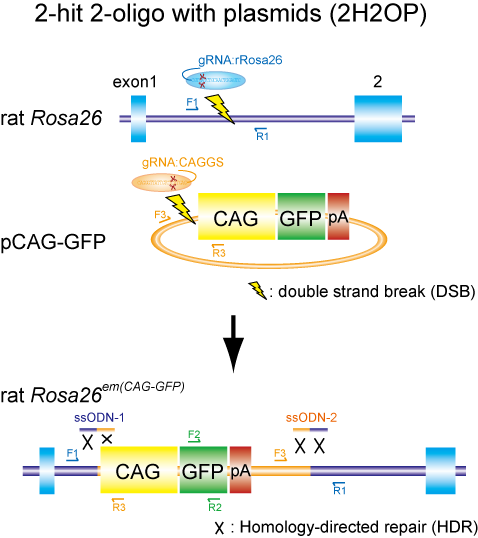
Fig.1 Schematic representation of the two-hit two-oligo with plasmid (2H2OP) method. This method enable efficient KI of plasmid DNAs by using single-stranded oligodeoxynucleotides (ssODNs) as ‘paste’.

Fig.2 GFP knock-in rats generated by 2H2OP method. Some of the pups express GFP at a high level in the body.
To learn more about this research, please view the full research report entitled " ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes " at this page of the Nature Communications website.
Related links
