The chemistry lab inside cells
A research team led by Osaka University has identified the protein used by the body to convert amino acids into the enzyme cofactor cysteine tryptophylquinone, which may lead to the development of novel biological catalysts
Investigators from the Institute of Scientific and Industrial Research at Osaka University, together with Hiroshima Institute of Technology, have announced the discovery of a new protein that allows an organism to conduct an initial and essential step in converting amino acid residues on a crosslinked polypeptide into an enzyme cofactor. This research may lead to a better understanding of the biochemistry underlying catalysis in cells.
Every living cell is constantly pulsing with an array of biochemical reactions. The rates of these reactions are controlled by special proteins called enzymes, which catalyze specific processes that would otherwise take much longer. A number of enzymes require specialized molecules called “cofactors,” which can help shuttle electrons back and forth during oxidation-reduction reactions. But these cofactors themselves must be produced by the organisms, and often require the assistance of previously existing proteins.
Now, a team of scientists at Osaka University has identified a novel protein called QhpG that is essential for the biogenesis of the enzyme cofactor cysteine tryptophylquinone (CTQ). By analyzing the mass of the reaction products and determining its crystal structure, they were able to deduce the catalytic function of QhpG, which is adding two hydroxyl groups to a specific tryptophan residue within an active-site subunit QhpC of quinoheme protein amine dehydrogenase, the bacterial enzyme catalyzing the oxidation of various primary amines. The resulting dihydroxylated tryptophan and an adjacent cysteine residue are finally converted to cofactor CTQ.
However, the action of QhpG is somewhat unusual compared with other protein-modifying enzymes in that it reacts with the tryptophan residue on the QhC triply crosslinked by another enzyme QhpD in a process call post-translation modification. Tryptophan, which naturally contains rings with conjugated bonds, needs the fewest changes to become a quinone cofactor. “Although several enzymes are known to contain a quinone cofactor derived from a tryptophan residue, the mechanism involved in post-translational modification, as well as the structures of the enzymes involved in their biogenesis, remains poorly understood,” lead author Toshinori Oozeki says.
The proteins were obtained by introducing plasmids with the corresponding genes into E. coli bacteria and made into crystals. X-ray diffraction data of the crystal can determine the QhpG protein structure. The team then used computer software to simulate the docking of the target molecules, the triply crosslinked polypeptide QhpC, based on the crystal structure they found for QhpG. The two post-translational modifications of QhpC are successively carried out in the modification enzyme complex QhpD-QhpG. “Our findings can be applied to development of novel bioactive peptides using enzymes that modify amino acids,” senior author Toshihide Okajima says. Some of these applications include creating new enzymes for the bioremediation of toxic chemicals.
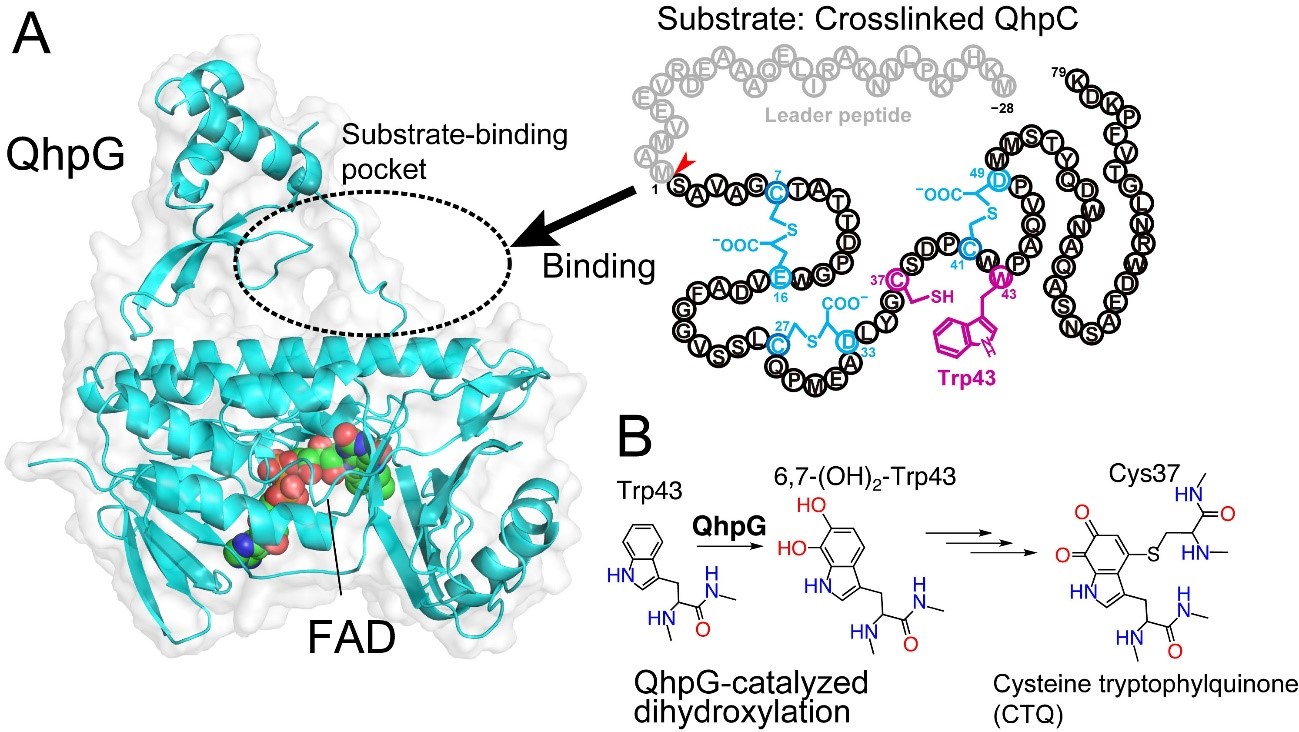
Fig. 1. (A) X-ray crystal structure of QhpG and schematic of crosslinked QhpC. The substrate QhpC is bound to the pocket formed by the catalytic domain, which includes the FAD cofactor and the small domain. (B) QhpG-catalyzed dihydroxylation reaction.
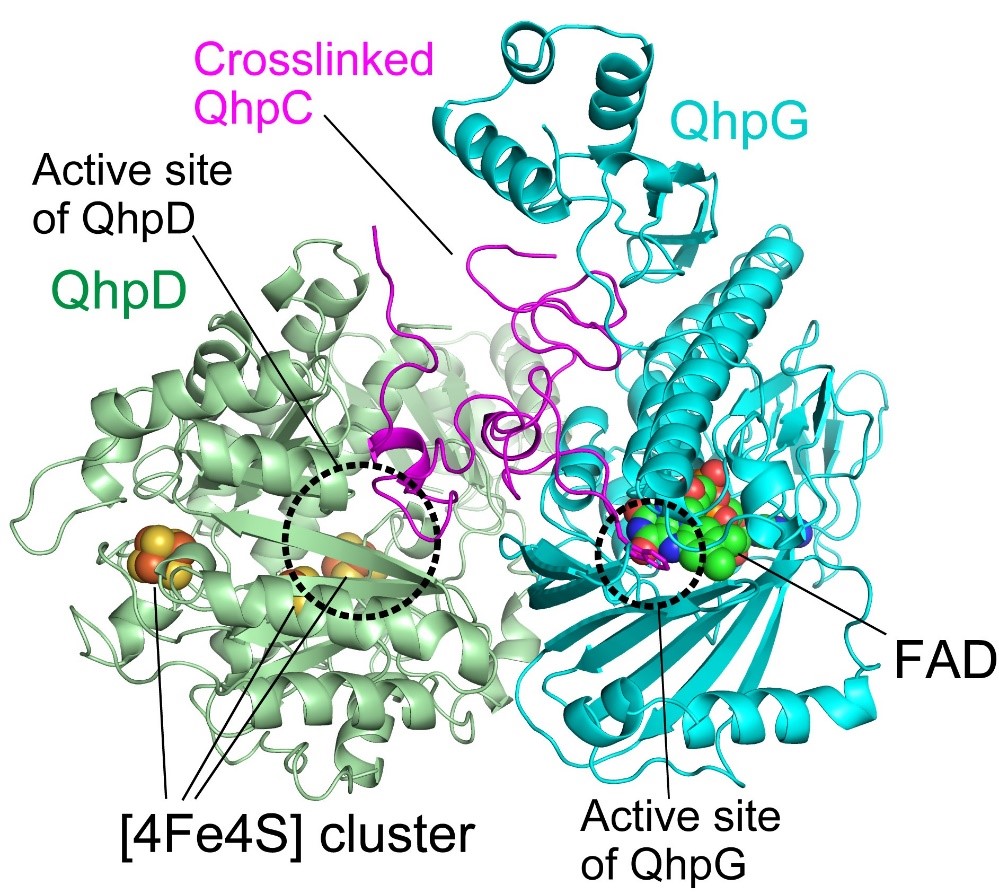
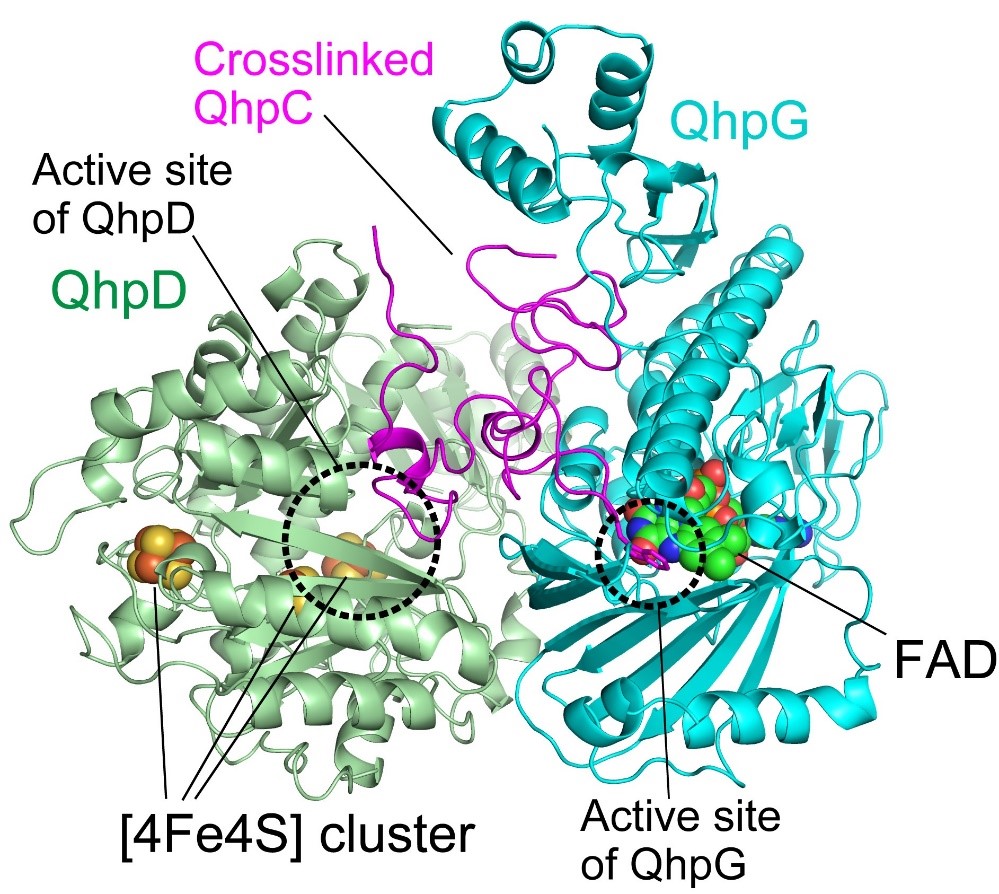
Fig. 2. Model of QhpC/QhpD/QhpG ternary complex. In this complex, QhpC is triply crosslinked at the active-site pocket of QhpD, along with [4Fe4S] clusters. Subsequently, a specific Trp residue of the crosslinked QhpC becomes dihydroxylated at the active site of QhpG close to the FAD. The complex formation thus enables efficient and sequential posttranslational modifications.
The article, “Functional and structural characterization of a flavoprotein monooxygenase essential for biogenesis of tryptophylquinone cofactor,” was published in Nature Communications at DOI: https://doi.org/10.1038/s41467-021-21200-9.
