Up to our NECs in it: in-depth genomic analysis of a rare carcinoma
Researchers from Osaka University identify several key biological processes underlying the development of neuroendocrine carcinoma of the gastrointestinal system, a rare and very lethal form of cancer
Researchers from Osaka University investigated a highly lethal form of cancer in a large group of patients, and identified the genetic culprits that lead to deadly outcomes.
In a study published in Cancer Discovery, a journal of the American Association for Cancer Research, researchers from Osaka University have thoroughly revealed the genetic basis of the pathogenesis of neuroendocrine carcinoma (NEC) of the gastrointestinal system, a rare and deadly cancer that is highly resistant to treatment.
NECs are cancers that originate in most epithelial organs of the body and most often in the digestive system, typically the pancreas. In addition to NEC being a rare cancer, patients with NEC do not often undergo surgery, so tissue samples that can be used for research purposes are hard to find.
“Because of this, the genetic changes that contribute to the development of this cancer have remained largely unexplored until now,” says lead author of the study Shinichi Yachida. “By taking part in an international collaboration, we were able to conduct a comprehensive genomic analysis of a relatively large number of cases.”
To investigate the genetic basis of NEC in these patients, the researchers performed a variety of analyses, including whole-genome sequencing, transcriptome sequencing, DNA methylation analysis, assay of transposase accessible chromatin sequencing, and whole-exome sequencing on tissue samples taken from patients.
“The results provided us with an unprecedented level of insight into the pathogenic mechanisms of NEC,” states Tatsuhiro Shibata, senior author. “For example, we found that pancreatic NECs are genetically distinct from pancreatic neuroendocrine tumors and may not be involved in the same carcinogenesis.”
Structural variants, in which part of a chromosome is inserted, deleted, or inverted, were far more common in nonpancreatic NECs than they were in pancreatic NECs. In addition, pancreatic NECs could be classified into two different groups (“ductal-type” and “acinar-type”) based on their genomic features. Intriguingly, the researchers also identified unusually high levels of methylation on the promoter of a transcription factor associated with NEC; a previously unknown genetic event where two genes became fused together, creating a new hybrid gene that disrupted cellular function; and deletion of an RNA splicing factor that has not previously been linked to NEC.
“Our study suggests that different types of NECs arise due to very different sets of driver mutations and genomic changes, which could have important implications for patients’ treatment,” explains Yachida.
Given that this is one of the most comprehensive studies of this cancer to date, it is likely that the findings will help develop new, more effective treatments for affected patients. Existing drugs could be used in some of these patients to specifically target the genomic changes leading to disease, and the targets defined in this study could even promote new drug discovery.
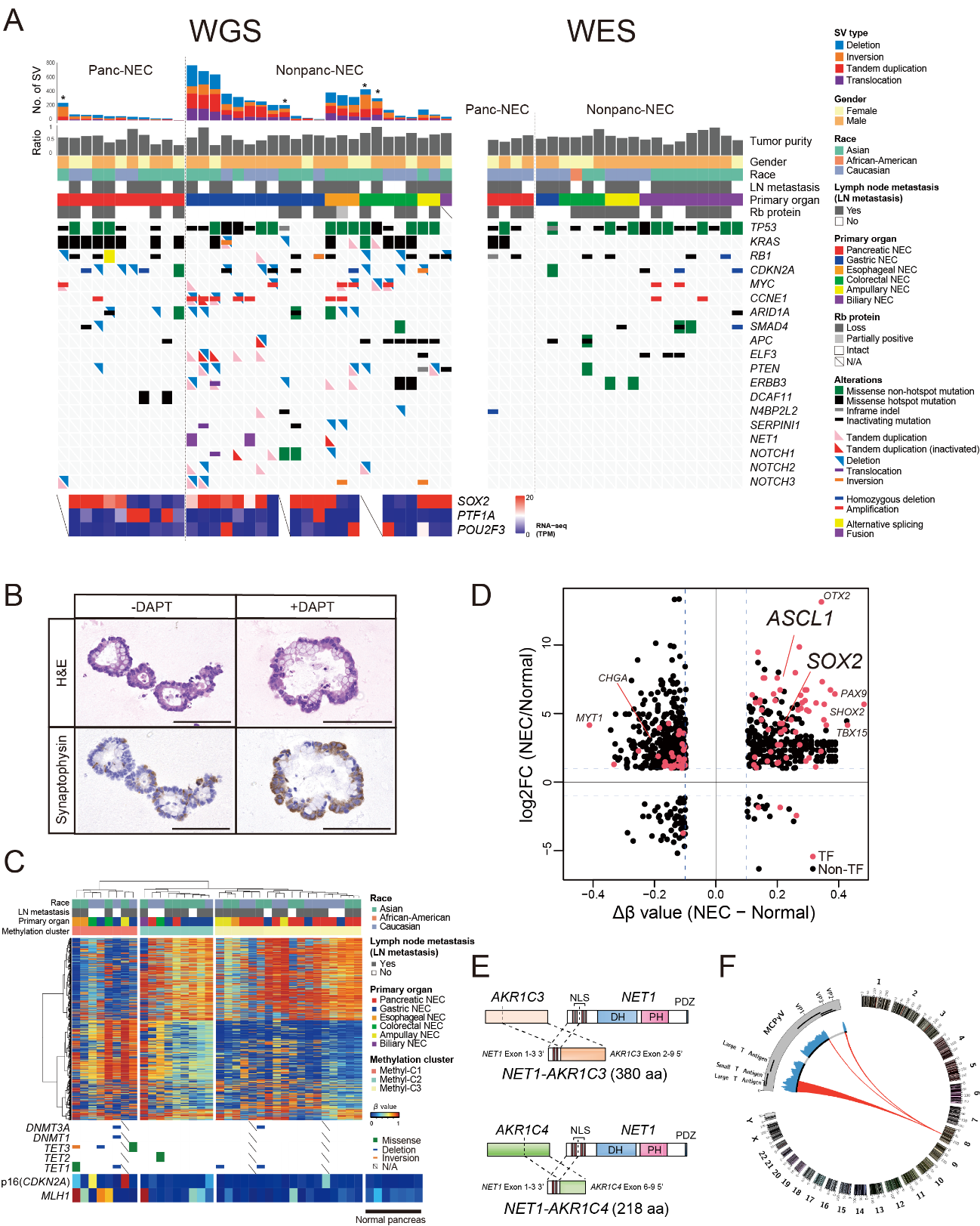
Figure 1. Genomic alterations in gastrointestinal system neuroendocrine carcinomas (GIS-NECs).
A, Landscape of genomic alterations in samples from patients with GIS-NEC. The left oncoplot shows whole-genome sequencing (WGS) data and representative gene expression data obtained from frozen samples. The cases are arranged from left to right in descending order of the number of structural variants in each primary organ. Asterisks, organoid samples. The right oncoplot shows whole-exome sequencing data from a different subset of patients than those for whom WGS data were available. B, H&E staining and synaptophysin immunolabeling of TP53 and RB1 double knockout (TR-KO) organoids before and after Notch signaling was blocked with a γ-secretase inhibitor (DAPT). The number of synaptophysin-positive cells was increased by treatment with the Notch inhibitor. Scale bar, 100 µM. C, Unsupervised hierarchical cluster analysis using 2,000 high variant probes for DNA methylation in GIS-NECs. D, Integration of RNA-seq and DNA methylation array data comparing GIS-NECs with normal tissues. RNA-seq data were filtered using significant differentially expressed genes (DEGs) (abs [log2FC]) ≥ 1 with significant false discovery rate values (< 0.05). DNA methylation assay data were filtered using differentially methylated regions (DMR) (abs [Db value] ≥ 0.1) with significant adjPvals (< 0.05). The area of the figure showing high gene expression level and hypermethylation contains 199 DMRs, of which 39 (19.6%) are transcription factors (TFs, red dots), including SOX2 and ASCL1. In contrast, the area of the figure showing high gene expression level and hypomethylation contains 424 DMRs, of which 28 (6.6%) are TFs. CHGA, chromogranin A. E, Schematic of NET-AKR fusion genes detected in two gastric NECs. The neuroepithelioma transforming 1 (NET1) gene is a specific guanine nucleotide exchange factor for RhoA. Both aldo-keto reductase family 1 members C3 (AKR1C3) and C4 (AKR1C4) are reductase enzymes that play critical roles in the biotransformation of endogenous substrates such as steroids. The chimeric genes demonstrate in-frame fusion of the NET1 amino terminus (exons 1–3) and the AKR1C3 carboxyl terminus (exons 2–9) or the AKR1C4 carboxyl terminus (exons 6–9). NLS, nuclear localization signal; DH, Dbl homology; PH, pleckstrin homology; PDZ, post-synaptic density 95; aa, amino acids. F, Gastric NEC with the Merkel cell polyomavirus (MCPyV) [Case NE002]. The read depth along the polyomavirus genome is shown in blue, and read pairs bridging the polyomavirus genome and the integration site on chromosome 8 are indicated by red lines. Polyomavirus genes are indicated by Large T Antigen, Small T Antigen, VP1, VP2, and VP3. (credit: ©2021 Shinichi Yachida et al., Cancer Discovery)
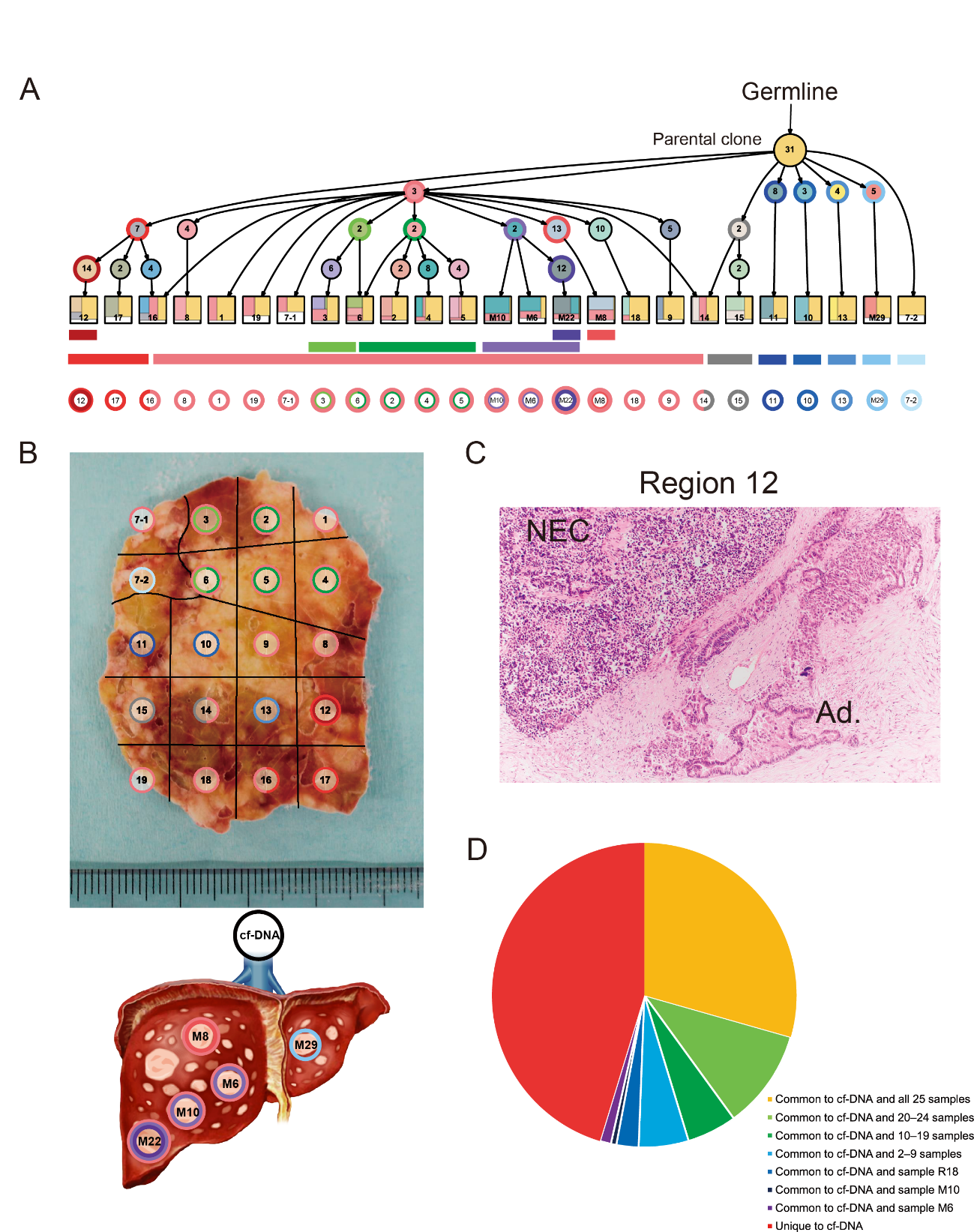
Figure 2. Geographic mapping of subclones based on multi-region whole-exome sequencing and proposed clonal evolution of a pancreatic neuroendocrine carcinoma (Panc-NEC) in an autopsied patient.
A, Proposed clonal evolution model according to the evolutional lineage tree based on the variant allele frequency of the mutations (LICHeE) in 20 primary regions and five liver metastases. The numbers inside the circles indicate mutations used by LICHeE to infer the subclonal structure. The colors in each subdivision describe the mutation groups characterizing cells in this subpopulation. The numbers and colors inside the squares indicate the region numbers shown in B and composition of subpopulations, respectively. B, Macroscopic view of a section of the largest part of the primary Panc-NEC. Different areas are marked with colors corresponding to the predicted subclones based on the evolutional lineage tree. C, Microscopic view of region 12 of the primary Panc-NEC (H&E staining). Copy number analysis demonstrated that whole genome duplication occurred in only the adjacent regions 12, 16, and 17. An adenocarcinoma component was observed together with NEC only in region 12. NEC, NEC component; Ad., adenocarcinoma component. D, Pie chart showing the relationship between mutations detected in plasma cell free DNA and in tissue samples (20 primary regions and five liver metastases).
(credit: ©2021 Shinichi Yachida et al., Cancer Discovery)
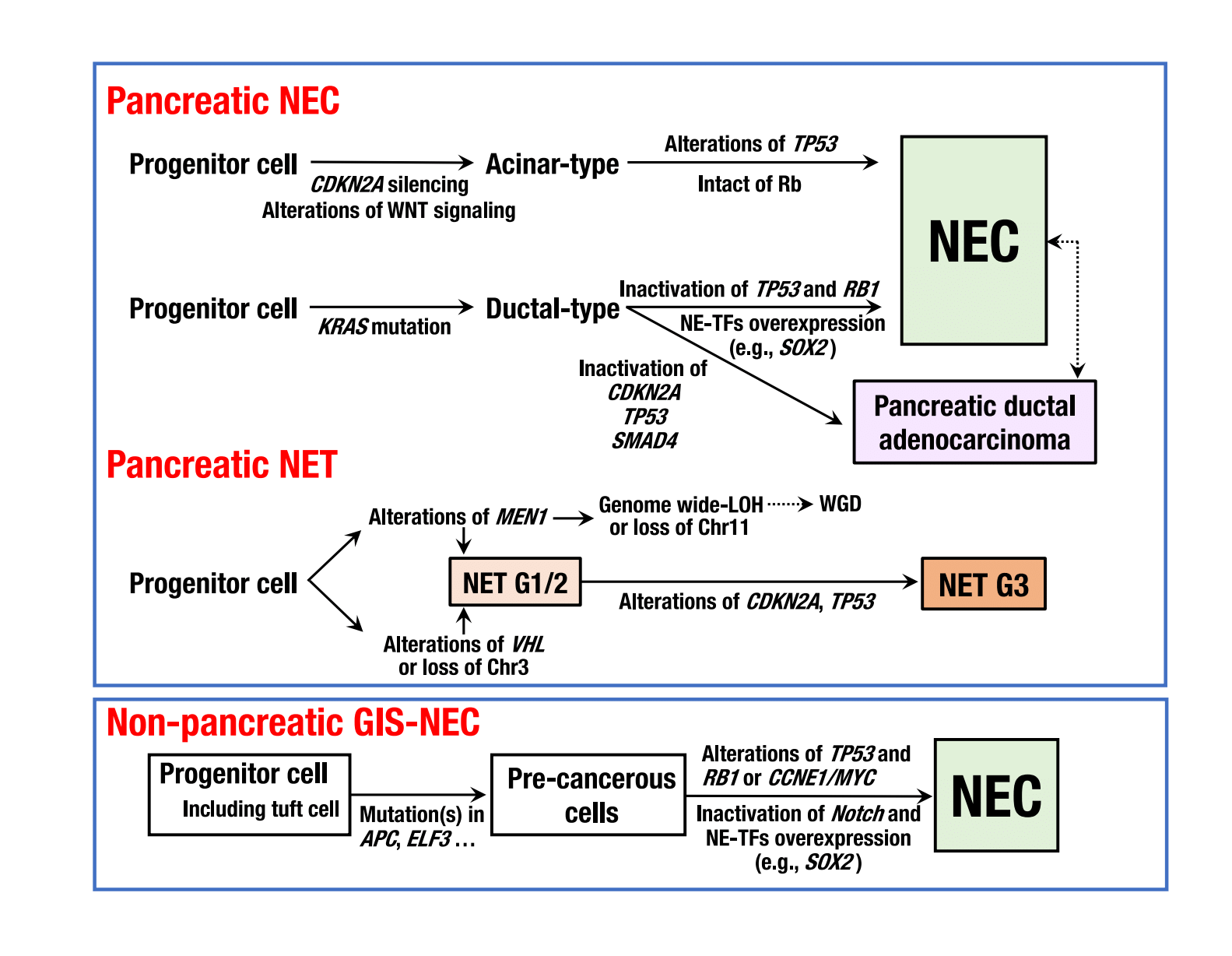
Figure 3. Schematic diagram of genomic alterations involved in the genesis of pancreatic neuroendocrine carcinoma, pancreatic neuroendocrine tumor, and non-pancreatic neuroendocrine carcinoma of the gastrointestinal system.
NE-TFs, neuroendocrine differentiation transcription factors. (credit: ©2021 Shinichi Yachida et al., Cancer Discovery)
The article, “Comprehensive genomic profiling of neuroendocrine carcinomas of the gastrointestinal system,” was published in Cancer Discovery at DOI: https://doi.org/10.1158/2159-8290.CD-21-0669.
