When FRETing over cancer biomarkers won't work, focus on blinking instead
Researchers from Osaka University used a photostabilizer to access otherwise hidden chemical details of DNA conformational dynamics, and to detect a cancer RNA biomarker at ultrahigh sensitivity
Fluorescence spectroscopy is indispensable in biomedical diagnostics. One can think of turning on fluorescence as turning on a flashlight in a dark room. A diagnostic assay can be designed to label, for example, a specific molecule of DNA with a fluorescent probe. If that specific molecule of DNA is present, you see fluorescence or a change in the fluorescence.
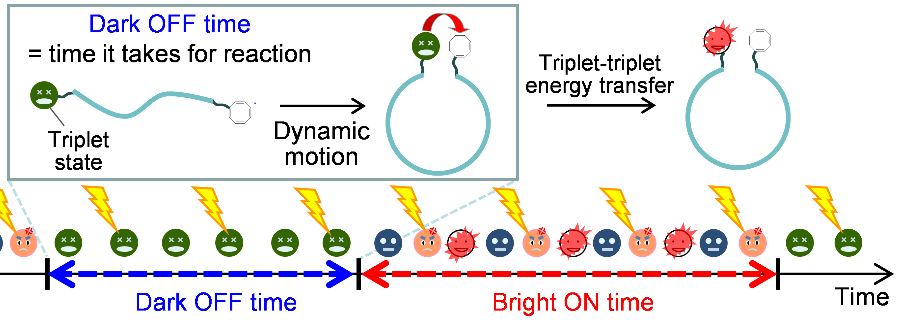
Sometimes an otherwise fluorescent molecule stops emitting light for a brief period of time. This is called fluorescence blinking, which can make it difficult to detect biomolecules at the ultralow concentrations necessary for disease diagnostics. A way to simultaneously decrease blinking for diagnostics and extract useful biochemical information from the blinking for basic research would be the best of both worlds.
In a study recently published in Angewandte Chemie, researchers from Osaka University used a well-known molecule abbreviated as COT—a photostabilizer—to modulate fluorescence blinking in biochemical assays. The researchers used COT to probe the architecture of DNA molecules and to detect a cancer RNA biomarker at ultralow concentrations.
"COT suppresses fluorescence blinking, and so increases fluorescence, by coming into physical contact with the fluorophore," explains Jie Xu, lead author. "In contrast, modulating emission by a widely-used technique known as fluorescence resonance energy transfer, FRET, works over only much longer distances—in the region of 1 to 10 nanometers—and only on a nanosecond timescale."
The researchers first tested their setup on double-stranded DNA containing an internal spacer. When COT was on one end of the spacer and the fluorophore at the other end, there was more fluorescence than when COT was not present. However, fluorescence blinking wasn't eliminated entirely. The researchers exploited this fact by testing how the chemical architecture of the spacer modulates blinking.
"Increasing spacer length and increasing pi-stacking interactions—noncovalent interactions between aromatic rings—in the spacer increased the fluorophore's time in the ‘off’ state," says Kiyohiko Kawai, senior author. "FRET can't provide information on biomolecular dynamics over these subnanometer distances."
The researchers next detected ultrasmall concentrations of an RNA molecule that is a biomarker for many cancers. They first affixed a fluorescent probe containing COT to a glass slide. The probe was designed such that binding to the RNA biomarker would increase fluorescence from the probe.
"Binding to the target RNA decreased the probe's time in the off state by half," says Xu. "This provides a clear means to detect a cancer biomarker."
Detecting a disease-pertinent biomolecule at ultralow concentrations, as made possible with this technique, can be a way to diagnose a disease in its early stages and facilitate treatment. Furthermore, many fundamental biochemical research studies are feasible now that researchers can probe molecular motions on the subnanometer scale and over broad timescales.
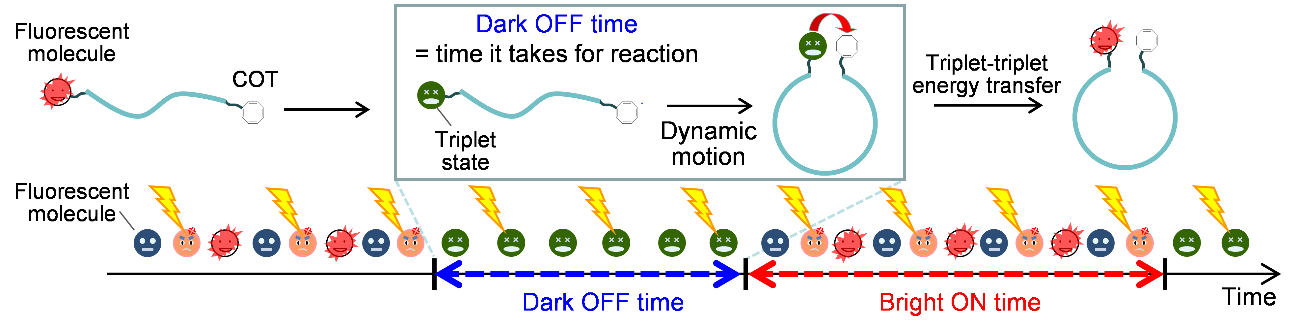
Figure 1 Schematic representation of fluorescence blinking controlled by triplet formation and triplet-triplet energy transfer.
The article, "Control of triplet blinking using cyclooctatetraene to access the dynamics of biomolecules at the single-molecule level," was published in Angewandte Chemie at DOI: https://doi.org/10.1002/anie.202101606.
