High-speed atomic force microscopy clarifies how enzyme repairs DNA damage
A group of researchers from Osaka University, Nagoya University, and Nara Institute of Science and Technology has succeeded in observing MRE11/RAD50/NBS1 complex (MRN), an enzyme to repair damage in human DNA.
A genome is a complete set of instructions for making an organism and contains the master blueprint for all cellular structures and activities for the lifetime of the cell or organism. If a genome is damaged by carcinogens or exposure to radiation, genetic information is lost and cells become cancerous or die.
Human MRN plays an important role in the mechanisms for repairing DNA damage. DNA damage accumulation may exacerbate the aging process and increase the risk of cancer development. Cellular DNA repair machinery, such as the MRN complex, is also necessary for genome editing in cells and organisms. This group observed the molecular structure of MRN with high-speed atomic force microscopy (HS-AFM) developed in Japan.
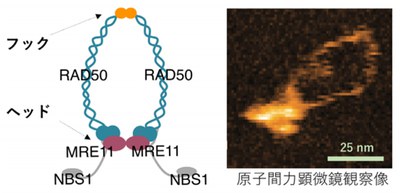
MRN is composed of two MRE11 subunits, two RAD50 units, and two NBS1 subunits. It was thought that MRN opened at the zinc hook of RAD50 and that the two arms which protrude from the large globular structure of MRN were used for bridging DNA ends in DNA double strand break repair.
In this study, however, imaging of the human MRE11/RAD50 complex by high-speed AFM showed that the RAD50-coiled coil arms were consistently bridged by the dimerized hooks, while the MRE11/RAD50 ring opened by disconnecting the head domains. In the head-open structure, two MRE11/RAD50 heterodimers were linked only at the hook and the two globular domains were separated.
This group examined the molecular structures of yeast and E-coli, finding that the overall architecture of the yeast MRE11/RAD50 was remarkably similar to that of the human MRE11/RAD50. These architectural features are conserved in the yeast and bacterial MRE11/RAD50 complexes. It is thought that movement of the coiled coil arms is an intrinsic feature of MRN.
In addition, yeast strains harboring the chimeric MRE11/RAD50 complex containing the SMC hinge of bacterial condensin MukB, instead of the RAD50 hook, properly functioned in DNA repair. This means that MRN opens at the head. Thus, this group proposed that the basic role of the RAD50 hook is similar to that of the SMC hinge, which serves as a rather stable dimerization interface.
Understanding the shape and movement of enzymes is important fundamental research in understanding functions of enzymes and how to control their activity. If it becomes possible to efficiently control MRN in cells, it will be useful for applied research regarding radiotherapy, anti-cancer treatment, and genome editing.
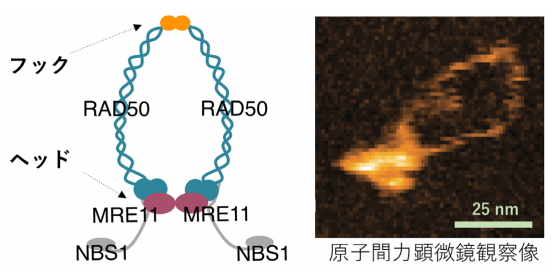
Figure 1
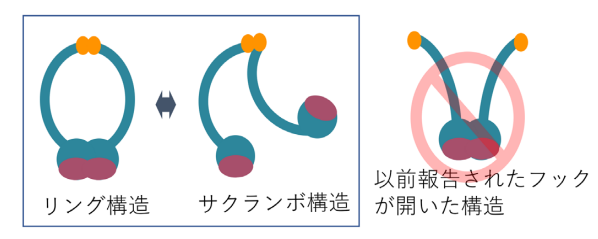
Figure 2
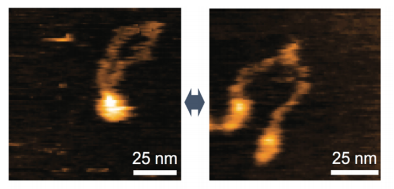
Figure 3
The article, "Rad50 zinc hook functions as a constitutive dimerization module interchangeable with SMC hinge," was published in Nature Communications at DOI: https://doi.org/10.1038/s41467-019-14025-0.
