Keeping Chromosomes in Check: A New Role for Heterochromatin
A team led by researchers at Osaka University finds that extra-tight packaging of genomic material helps prevent large chromosomal rearrangements that can lead to cancer
Although many people are aware that chromosomal damage and shortening contribute to the aging process, understanding how chromosomal defects occur is about more than just finding a way to turn back the clock. Large changes in the structure of chromosomes, known as gross chromosomal rearrangements, can result in cell death or genetic diseases such as cancer.
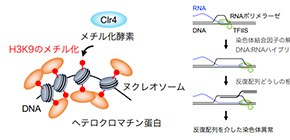
Heterochromatin is a more densely-coiled version of chromatin, the mass of DNA and proteins that forms chromosomes. A unique region of a chromosome called the centromere is vital for correct segregation of chromosomes during cell division. While researchers have long known that centromeres are composed of heterochromatin, why the centromere is so tightly packed and how this helps stabilize the region has remained elusive—until now.
In a recent paper published in Communications Biology , a research team led by Osaka University has uncovered the role of heterochromatin in maintaining chromosomal integrity.
Centromeric regions contain a large number of short, repeated DNA sequences. These repeats make the centromere especially susceptible to breakage and rearrangements, often causing one entire arm of a chromosome to be lost while the other arm is duplicated, forming structures called isochromosomes. But the team from Osaka University found that a specific feature of heterochromatin, histone H3 lysine 9 (H3K9) methylation, suppresses gross chromosomal rearrangements caused by centromere repeats.
“Deletion of Clr4, the protein responsible for H3K9 methylation in model organism Schizosaccharomyces pombe , caused an increase in the formation of isochromosomes with breakpoints located in the centromere repeats,” says lead author Akiko Okita, suggesting that methylation helps prevent rearrangements.
However, further investigation revealed that the mechanism was even more intricate than first thought.
Heterochromatin silences the action of RNA polymerase II, an enzyme responsible for copying DNA into RNA transcripts. Unexpectedly, complete silencing of RNA transcription did not seem to be necessary to suppress gross chromosomal rearrangements. Transcription factor Tfs1/TFIIS is needed to restart RNA polymerase II if it backtracks along the already-copied DNA sequence. Intriguingly, the researchers found that deletion of Tfs1/TFIIS was sufficient to bypass the need for Clr4 in the suppression of gross chromosomal rearrangements, and that RNA transcription levels were largely unaffected by the Tfs1/TFIIS deletion.
“The results showed that repression of Tfs1/TFIIS-dependent ‘persistent’ transcription of centromere repeats is the key role of heterochromatin in the suppression of gross chromosomal rearrangements,” explains corresponding author Takuro Nakagawa. Essentially, heterochromatin stops the repeats from being copied and used in the formation of gross chromosomal rearrangements.
“We predict that our findings will help develop methods of securing genome integrity by manipulating chromatin status rather than by changing the DNA sequence,” says Dr Nakagawa. “This would be a huge accomplishment because the ability to suppress gross chromosomal rearrangements is integral to the prevention of diseases arising from chromosomal instability.”

Fig. 1. Heterochromatin suppresses gross chromosomal rearrangements mediated by centromere repeats.
(credit: Osaka University)

Fig. 2. Loss of Clr4/Suv39 histone H3K9 methyltransferase increases the spontaneous rate of gross chromosomal rearrangements.
(credit: Osaka University)
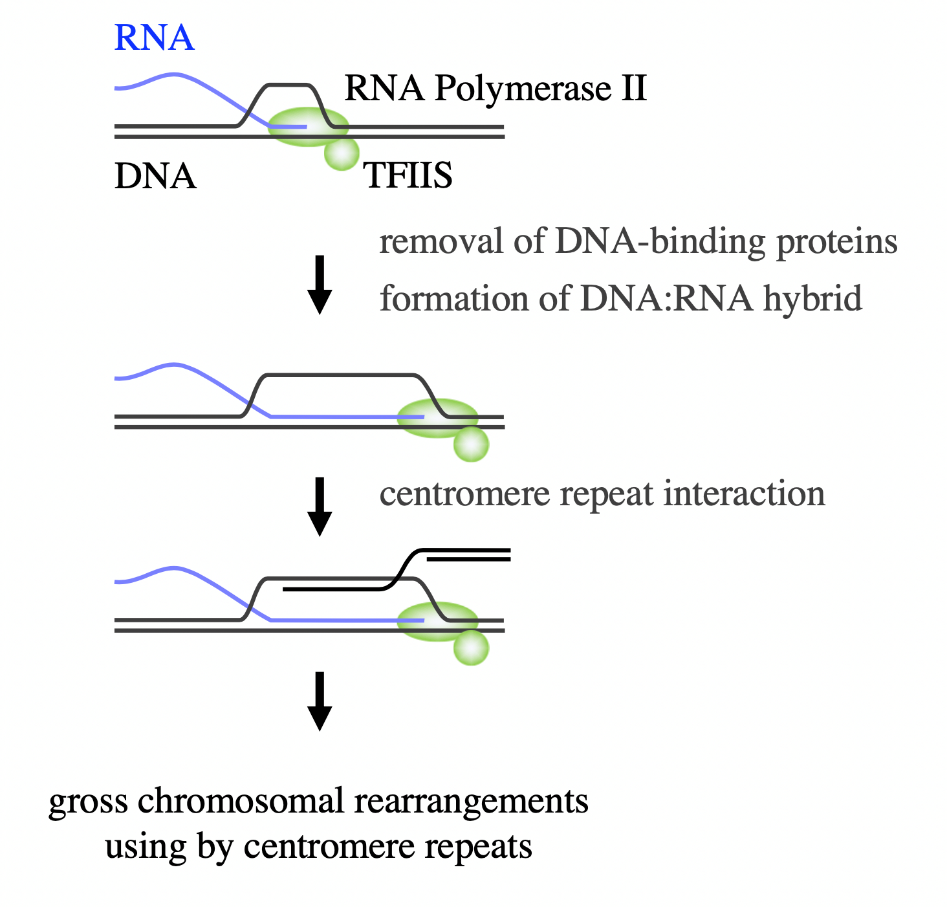
Fig. 3. Model outlining how Tfs1/TFIIS-dependent “persistent” transcription causes gross chromosomal rearrangements mediated by centromere repeats.
(credit: Osaka University)
The article, “Heterochromatin suppresses gross chromosomal rearrangements at centromeres by repressing Tfs1/TFIIS-dependent transcription,” was published in Communications Biology at DOI: https://doi.org/10.1038/s42003-018-0251-z .
Related links
