Artificial Fluorescent Membrane Lipid Shows Active Role in Living Cells
Osaka University-led international research collaboration develops synthetic fluorescent mimics of biological membrane lipid that can be monitored in living cells to reveal dynamic behavior
Biological membranes, such as those surrounding animal cells, are made up of lipids and proteins. Because these molecules do not usually mix well, they are distributed within different regions of the membrane. This segregation is achieved in a number of ways, including the formation of domains based on particular lipids such as cholesterol or sphingomyelin (SM). These two lipids are required for the generation of cholesterol-dependent raft domains, which are necessary for signaling within the plasma membrane. However, it was not clear how SMs interacted with other molecules of raft domains, mainly because of the lack of a suitable synthetic probe of SM. Now, research led by Osaka University in collaboration with JST ERATO Lipid Active Structure Project has developed new fluorescent synthetic molecules (analogs) that structurally mimic SMs and can be studied in live cells. The study was reported in J Cell Biol.
Existing fluorescent SM analogs behave differently from their fully functional natural counterparts. For example, they usually separate into a different type of fluid phase from that seen in living membranes. Moreover, those synthetic analogs that do split into the correct fluid phase produce a weak fluorescent signal, quickly lose their pigment, or sometimes need to be excited by UV light.
Researchers at Osaka University overcame these limitations with fluorescent SM analogs by joining several fluorescent chemical compounds (fluorophores) that were highly hydrophilic to the hydrophobic lipid part (mainly acyl chains) of the synthetic molecule. “We took care to ensure that the positive charge of the headgroup was maintained by not modifying its lipid part,” co-first author Masanao Kinoshita says. “This was achieved by keeping the fluorescent compounds away from the headgroup using a long linker component."
After confirming that the synthetic molecules behaved similarly to natural SM by using simple model membranes, the team next used highly sensitive single-molecule imaging to monitor the role of SMs in living cell membranes.
“We observed interactions of the SM analogs with each other and with CD59, which is a type of lipid receptor that is commonly used to link proteins to the plasma membrane,” corresponding author Nobuaki Matsumori says. “These interactions were shown to sometimes require the presence of cholesterol as well as an alcohol component of SMs.”
Further analysis revealed the dynamic behavior of SMs as they rapidly associated and dissociated from raft domains involving different formations of CD59 and with the plasma membrane. These findings may help in modifying future molecular interactions such as increasing their rate or complexity.
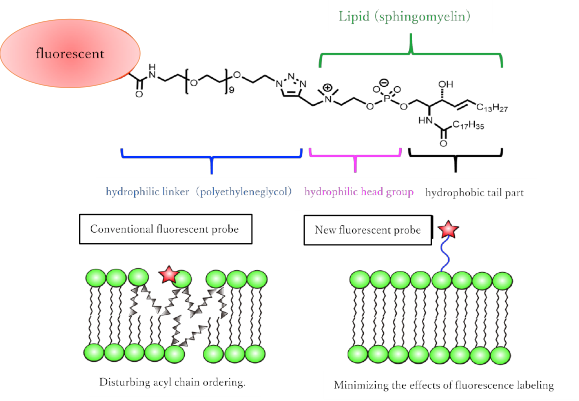
(Figure 1) (top) Newly synthesized sphingomyelin fluorescent analogue molecule. (Bottom) Differences from conventional fluorescent analog lipids. By using hydrophilic fluorescent molecule and linker, the same behavior as natural lipid can be reproduced.
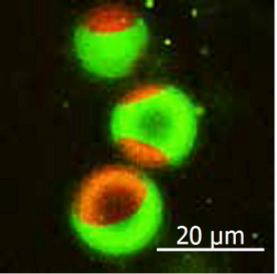
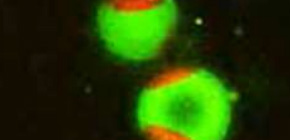
(Figure 2) The green fluorescent sphingomyelin molecule is localized in the raft-like region of the artificial membrane. The red area is a non-raft membrane area.
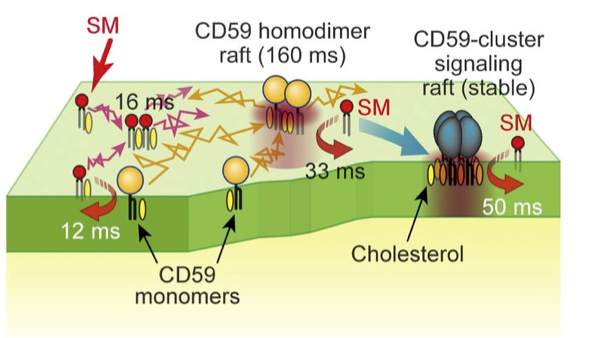
(Figure 3) Schematic diagram of transient binding of sphingomyelin to various forms of CD59, which was clarified in this study. (© 2017 Kinoshita M. et al. Journal of Cell Biology . VOL:216 NO:4 1183-1204. doi: 10.1083/jcb.201607086)
To learn more about this research, please view the full research report entitled "Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs" at this page of The Journal of Cell Biology.
Related link
Laboratory for Biomolecular Chemistry (Murata Lab), Graduate School of Science, Osaka University
http://www.chem.sci.osaka-u.ac.jp/lab/murata/english/index.html
