Proteins Recognize the Various Lengths of Fatty Acids Using a Water Cluster
This finding sheds light on molecular mechanism of a promiscuous lipid-protein interaction
Fatty acid-binding proteins (FABPs) are small proteins for cytosolic delivery of fatty acids (FAs), which are used as fuels for aerobic exercise and/or raw materials for inflammatory mediators. Assoc. Prof. Shigeru Matsuoka , Assoc. Prof. Shigeru Sugiyama , and Prof. Michio Murata at Osaka University revealed that human heart-type FABP (FABP3) identifies FAs not by exact matching but by “fuzzy recognition” of fundamental structural similarities among numerous FAs, where two different type of water clusters are involved in measuring the length of string-like molecule FAs. This finding sheds light on a molecular mechanism of a promiscuous lipid-protein interaction that would play key roles in FA-related diseases, such as metabolic syndrome and schizophrenia. The research was published online in the internationally leading chemistry journal, Angewandte Chemie International Edition, on December 9, 2014.
Abstract
Long-chain fatty acids (FAs) with low water solubility require fatty-acid-binding proteins (FABPs) to transport them from cytoplasm to the mitochondria for energy production. However, the precise mechanism by which these proteins recognize the various lengths of simple alkyl chains of FAs with similar high affinity remains unknown. To address this question, we employed a newly developed calorimetric method for comprehensively evaluating the affinity of FAs, sub-Angstrom X-ray crystallography to accurately determine their 3D structure, and energy calculations of the coexisting water molecules using the computer program WaterMap. Our results clearly showed that the heart-type FABP (FABP3) preferentially incorporates a U-shaped FA of C10–C18 using a lipid-compatible water cluster, and excludes longer FAs using a chain-length-limiting water cluster. These mechanisms could help us gain a general understanding of how proteins recognize diverse lipids with different chain lengths.
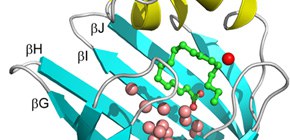
Figure 1: Structure of human heart-type fatty acid-binding protein (FABP3).
FABP3 has a clum-like shell structure which consists of a pair of five-stranded antiparallel β-sheets, called β-barrel (cyan), that surround a large water-filled cavity covered on the top by two short parallel α-helices (yellow). The cavity accommodates about 13 water molecules (pink and red spheres) and a fatty acid (green ball and stick model).
Figure 2: Isothermal titration calorimetry of fatty acid (FA) and dimyristoyl phosphatidylcholine (DMPC) liposomes.
a) FA (oleic acid) micelles; b) DMPC liposomes; and c) FA/DMPC (1/11 mol/mol) mixed liposomes were titrated into FABP3 solutions. Heat of binding was observed only when FA and DMPC were added as mixed liposomes.
Figure 3: Hydration site analysis of FABP3. Colour gradation of the hydration sites is based on the free energy relative to bulk water; stable sites are shown in green and unstable sites in red. a) The C10 FA-bound state of FABP3. Note that water molecules are segregated into two clusters with distinct stability. b) The C18 FA-bound state. The extended chain of C18 FA displaces unstable water molecules in Cluster 2. c) The C22 FA-bound state. The further extended chain of C22 FA breaks into stable water molecules in Cluster 1, which causes decrease in FA-FABP3 binding affinity.
To learn more about this research, please view the full research report entitled " Water-Mediated Recognition of Simple Alkyl Chains by Heart-Type Fatty Acid-Binding Protein " at this page of the Angewandte Chemie website.
Related Link
