Technology for quick 3D structure determination of membrane proteins developed
SACLA’s new technology accelerates designing new drugs
A joint group of researchers from Osaka University, The University of Tokyo, Japan Synchrotron Radiation Research Institute (JASRI), and S Pring-8 A ngstrom C ompact Free Electron La ser (SACLA), RIKEN SPring-8 Center, succeeded in developing technology for quickly determining membrane protein structures by using the SACLA at the XFEL facility, a world first.
Unlike water-soluble proteins, handling of membrane proteins is difficult, and many challenges must be overcome in order to visualize their molecular structures. In particular, with newly developed methods, serial femtosecond crystallography with X-ray free electron lasers, no membrane protein structures have been determined.
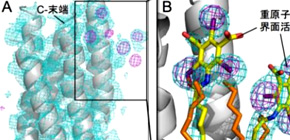
This research group developed a method for quickly determining membrane proteins by labeling membrane proteins through the synthesis of iodine-containing detergent and using SACLA’s experimental technology in combination. This group found that heavy-atom labeling of membrane proteins was available only by synthesizing an iodine-containing detergent -- iodine is a heavy atom with a high anomalous dispersion (SAD) -- and binding it with bacteriorhodopsin (bR) microcrystal, a membrane protein.
As a result of serial femtosecond X-ray crystallography, this group verified that 3D structures of membrane proteins could be determined by single-wavelength anomalous diffraction (SAD), single isomorphous replacement (SIR), or single isomorphous replacement with anomalous scattering (SIRAS), a world first.
This group also clarified that the iodine-containing detergent could also be used for G protein-coupled receptors (GPCR), which are an important membrane protein as a target for drug discovery, and demonstrated that this newly developed method for membrane protein structure determination was used for various membrane proteins.
This group’s achievement will promote membrane protein structure determination and understanding of biological phenomena at a molecular level. It is also hoped that this will lead to applications in medical fields including design and development of new drugs based on membrane protein structures, which are important targets for drug development.
Abstract
The 3D structure determination of biological macromolecules by X-ray crystallography suffers from a phase problem: to perform Fourier transformation to calculate real space density maps, both intensities and phases of structure factors are necessary; however, measured diffraction patterns give only intensities. Although serial femtosecond crystallography (SFX) using X-ray free electron lasers (XFELs) has been steadily developed since 2009, experimental phasing still remains challenging. Here, using 7.0-keV (1.771 Å) X-ray pulses from the SPring-8 Angstrom Compact Free Electron Laser (SACLA), iodine single-wavelength anomalous diffraction (SAD), single isomorphous replacement (SIR), and single isomorphous replacement with anomalous scattering (SIRAS) phasing were performed in an SFX regime for a model membrane protein bacteriorhodopsin (bR). The crystals grown in bicelles were derivatized with an iodine-labeled detergent heavy-atom additive 13a (HAD13a), which contains the magic triangle, I3C head group with three iodine atoms. The alkyl tail was essential for binding of the detergent to the surface of bR. Strong anomalous and isomorphous difference signals from HAD13a enabled successful phasing using reflections up to 2.1-Å resolution from only 3,000 and 4,000 indexed images from native and derivative crystals, respectively. When more images were merged, structure solution was possible with data truncated at 3.3-Å resolution, which is the lowest resolution among the reported cases of SFX phasing. Moreover, preliminary SFX experiment showed that HAD13a successfully derivatized the G protein-coupled A2a adenosine receptor crystallized in lipidic cubic phases. These results pave the way for de novo structure determination of membrane proteins, which often diffract poorly, even with the brightest XFEL beams.
Lecturer MIZOHATA Eiichi (Osaka University), Professor MURATA Michio (Osaka University), Group Director IWATA So (RIKEN) (from the left)
To learn more about this research, please view the full research report entitled “ Membrane protein structure determination by SAD, SIR,or SIRAS phasing in serial femtosecond crystallography using aniododetergent ” at this page of the Proceedings of the National Academy of Sciences of the United States of America (PNAS) website.
Related links
