Changes in genetic structure of yeast lead to disease-causing genomic instabilities
Researchers from The University of Osaka reveal that genetic changes following the loss of heterochromatin can lead to chromosomal rearrangements, potentially explaining the development of various diseases
Changes in genes have been linked to the development of different diseases for a while. However, it’s not exactly clear what the mechanisms, or the causes behind those specific genetic changes, are. Recent studies using fission yeast, which can act as an ideal model for human cells, have highlighted one possible mechanism linked to disease onset.
In a study recently published in Nucleic Acids Research, researchers from The University of Osaka discovered that the loss of heterochromatin can kickstart genetic changes, potentially resulting in the development of diseases like cancer.
The model showed that RNA-loops (R-loops) accumulate at clusters of repetitive DNA called pericentromeric repeats in response to a process called transcriptional pausing–backtracking–restart (PBR). These accumulated R-loops are then changed into Annealing-induced DNA-RNA-loops (ADR-loops), leading to gross chromosomal rearrangements (GCRs) at constricted parts of a chromosome.
“Previously, we showed that loss of Clr4, the H3K9me2/3 methyltransferase, or its regulatory protein Rik1, increased transcription and abnormal chromosome formation in fission yeast,” explains lead author, Ran Xu. “However, the molecular link between transcription dynamics and GCRs remains poorly defined.”
Heterochromatin forms at pericentromeric repeats. Previous research showed that heterochromatin could prevent GCRs at centromeres by blocking pericentromeric transcription. However, the present study expanded on past findings by providing insights into the mechanism by which GCRs are generated, including through pericentromeric transcription.
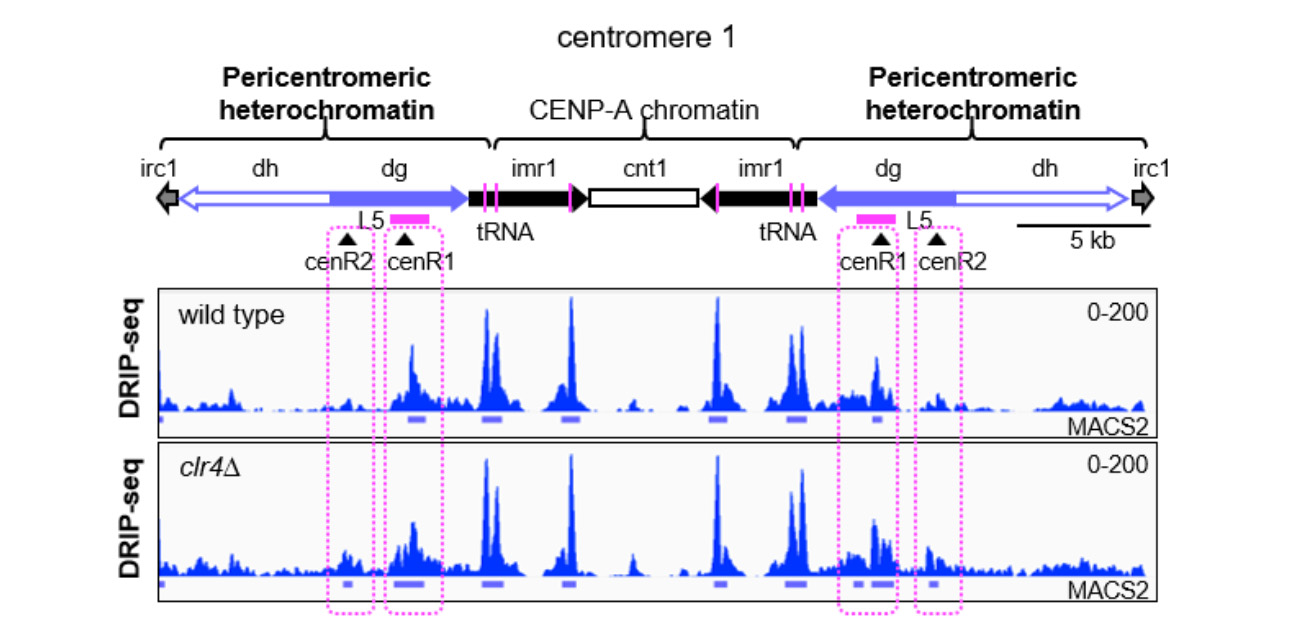
The researchers demonstrated that loss of Clr4 can spark an increase in R-loop levels at pericentromeric repeats. After overexpressing the enzyme RNase H1 in cells lacking the clr4 gene, the research team observed reductions in both R-loops and GCRs.
Further experiments highlighted the importance of Tfs1/TFIIS and Ubp3, which are necessary to restart transcription, in R-loop accumulation and GCRs. In cells lacking Clr4, a type of protein called Rad52 built up at pericentromeric repeats. This promoted the development of GCRs, and cells carrying a mutated version of this protein had fewer GCRs because single-strand annealing (SSA), a DNA repair process, was inhibited.
“These data suggest that, when heterochromatin is lost, transcriptional PBR cycles accumulate R-loops at pericentromeric repeats, and Rad52-dependent single-stand annealing converts R-loops into ADR-loops followed by Polδ-dependent break-induced replication (BIR), encouraging GCRs related to disease,” concluded Xu.
This study could have key insights for treating genetic diseases caused by GCRs, like cancer. Although further research is needed to translate these findings into human applications, drugs targeting Rad52 or other genes and proteins involved in GCR accumulation might emerge as key disease treatments.
Fig. 1
Caption: DNA-RNA Immunoprecipitation (DRIP)-Seq data showing accumulation of R-loops in the heterochromatin-deficient clr4∆ mutant.
Credit: 2026, Ran Xu et al., Transcriptional PBR cycles at pericentromeric repeats cause gross chromosomal rearrangements through Rad52-dependent ADR-loop formation, Nucleic Acids Research
Fig. 2
Caption: The Rad52 protein converts R-loops into ADR-loops, resulting in isochromosome formation.
Credit: 2026, Ran Xu et al., Transcriptional PBR cycles at pericentromeric repeats cause gross chromosomal rearrangements through Rad52-dependent ADR-loop formation, Nucleic Acids Research
Fig. 3
Caption: The model showing transcriptional PBR cycles accumulate R-loops, which are converted into ADR-loops by Rad52, resulting in gross chromosomal rearrangements.
Credit: 2026, Ran Xu et al., Transcriptional PBR cycles at pericentromeric repeats cause gross chromosomal rearrangements through Rad52-dependent ADR-loop formation, Nucleic Acids Research
Notes
The article, “Transcriptional PBR cycles at pericentromeric repeats cause gross chromosomal rearrangements through Rad52-dependent ADR-loop formation,” was published in Nucleic Acids Research at DOI: https://doi.org/10.1093/nar/gkaf1455
Links
- EurekAlert!
- AlphaGalileo
- Asia Research News
- Professor Nakagawa's Profile
- Laboratory of Molecular Genetics



