A NICER approach to genome editing
Researchers led by Osaka University develop a new gene modification technique known as NICER that significantly reduces off-target mutations in DNA
The gene editing technique CRISPR/Cas9 has allowed researchers to make precise and impactful changes to an organism’s DNA to fix mutations that cause genetic disease. However, the CRISPR/Cas9 method can also result in unintended DNA mutations that may have negative effects. Recently, researchers in Japan have developed a new gene editing technique that is as effective as CRISPR/Cas9 while significantly reducing these unintended mutations.
In a new study published in Nature Communications, researchers led by Osaka University introduced a novel technique called NICER, which is based on the creation of multiple small cuts in single DNA strands by an enzyme called a nickase.
Traditional CRISPR/Cas9 editing uses small pieces of genetic code called guide RNAs and an enzyme called Cas9. The guide RNAs target a specific section of the DNA and the Cas9 enzyme initiates a break in the double-stranded DNA structure at this location. This double-strand break is key for initiating changes to the DNA. However, cellular repair of double-strand breaks can lead to unintended DNA mutations, as well as the integration of exogenous DNA to the human genome, which raises safety concerns for clinical applications of CRISPR/Cas9 technology. To minimize these unintended mutations, the Osaka University-led research team investigated the use of Cas9 nickase, which creates single-strand breaks or “nicks” in DNA that are typically repaired without causing mutations.
“Each chromosome in the genome has a ‘homologous’ copy,” says lead author of the study Akiko Tomita. “Using the NICER technique, heterozygous mutations – in which a mutation appears in one chromosome but not its homologous copy – are repaired using the unmutated homologous chromosome as a template.”
For their initial experiments, the research team used human lymphoblast cells with a known heterozygous mutation in a gene called TK1. When these cells were treated with nickase to induce a single cut in the TK1 region, TK1 activity was recovered at a low rate. However, when the nickase induced multiple nicks in this region on both homologous chromosomes, gene correction efficiency was enhanced approximately seventeen-fold via activation of a cellular repair mechanism.
“Further genomic analysis showed that the NICER technique rarely induced off-target mutations,” says senior author Shinichiro Nakada. “We were also pleased to find that NICER was able to restore the expression of disease-causing genes in cells derived from genetic diseases involving compound heterozygous mutations.”
Because the NICER method does not involve DNA double-strand breaks or the use of exogenous DNA, this technique appears to be a safe alternative to conventional CRISPR/Cas9 methods. NICER may represent a novel approach for the treatment of genetic diseases caused by heterozygous mutations.
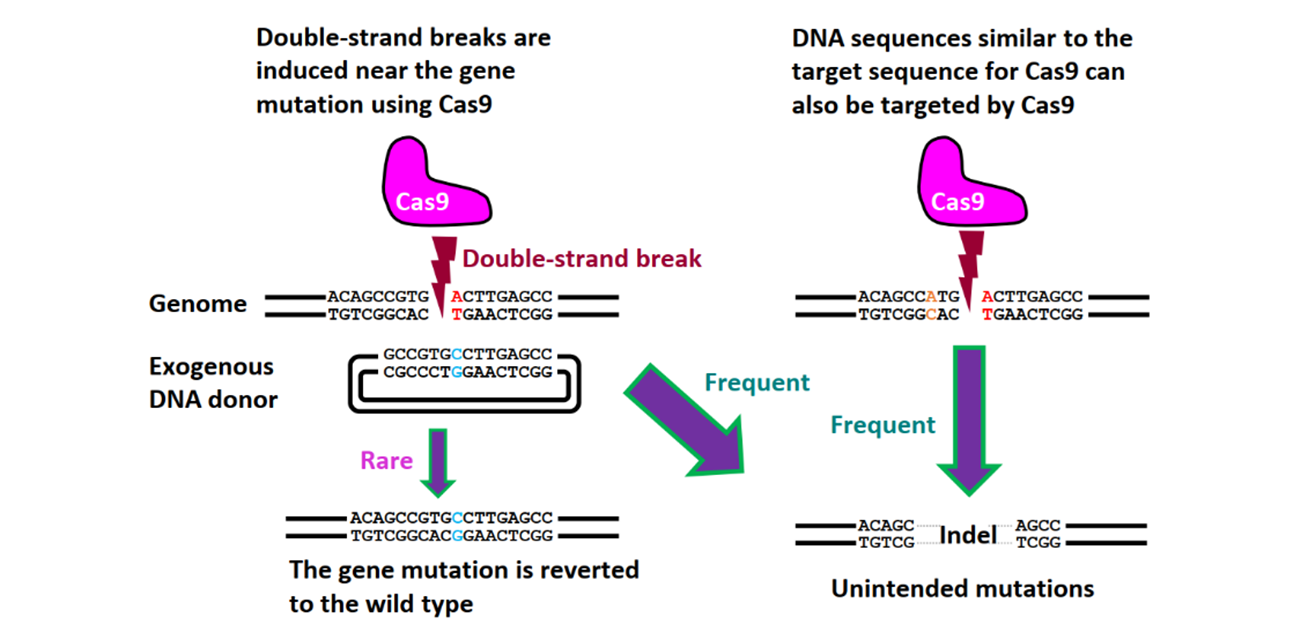
Fig. 1
Off-target mutations resulting from conventional genome editing methods. Due to the mutagenic nature of DNA double-strand break repair, even if the gene mutation is successfully corrected, there is a heightened risk of off-target mutations in areas distinct from the original target of CRISPR/Cas9.
Credit: S. Nakada
Fig. 2
Genome editing using the Cas9 mutant, nickase. Nickase induces single-strand breaks (nicks) instead of DNA double-strand breaks. Nicks are typically repaired without causing mutations. Hence, even if nicks appear at sites different from nickase's original target, new gene mutations are seldom seen at these locations. This approach allows for precise gene correction with minimal off-target mutations.
Credit: S. Nakada
Fig. 3
Schematic of the NICER gene editing method. A nick is made near the mutation (and is exclusive to the mutant allele). Merely introducing a nick close to the mutation instigates homologous recombination between homologous chromosomes but at a very low rate. However, when one or two additional nicks are made to both alleles, the gene correction efficiency through interhomolog homologous recombination sees a significant boost.
Credit: S. Nakada
The article, “Inducing multiple nicks promotes interhomolog homologous recombination to correct heterozygous mutations in somatic cells,” will be published in Nature Communications at DOI: https://doi.org/10.1038/s41467-023-41048-5.
