In situ extraction and detection of DNA is an im-pore-tant development
Researchers from Osaka University have used a three-dimensionally integrated nanopore to detect single DNA molecules extracted from individual cells
Being able to detect DNA from a single cell is important for the detection of diseases and genetic disorders. Measuring single DNA molecules has been possible for some time; however, directly detecting samples at the point of extraction with no need for subsequent steps has not. Now, researchers at SANKEN, Osaka University have demonstrated a method of releasing DNA at the point of measurement. Their findings are published in Small Methods.
Nanopores are very tiny holes that are found in biology or can be purpose engineered. There have been exciting advances in using nanopores as gateways that allow close monitoring as molecules pass through one by one. For example, the individual DNA bases passing through a pore have been identified allowing whole genome sequencing.
However, despite these remarkable steps in single molecule detection, it has been necessary to increase the concentration of DNA samples for successful measurement because there was no way of reliably getting the molecules to the measurement pore.
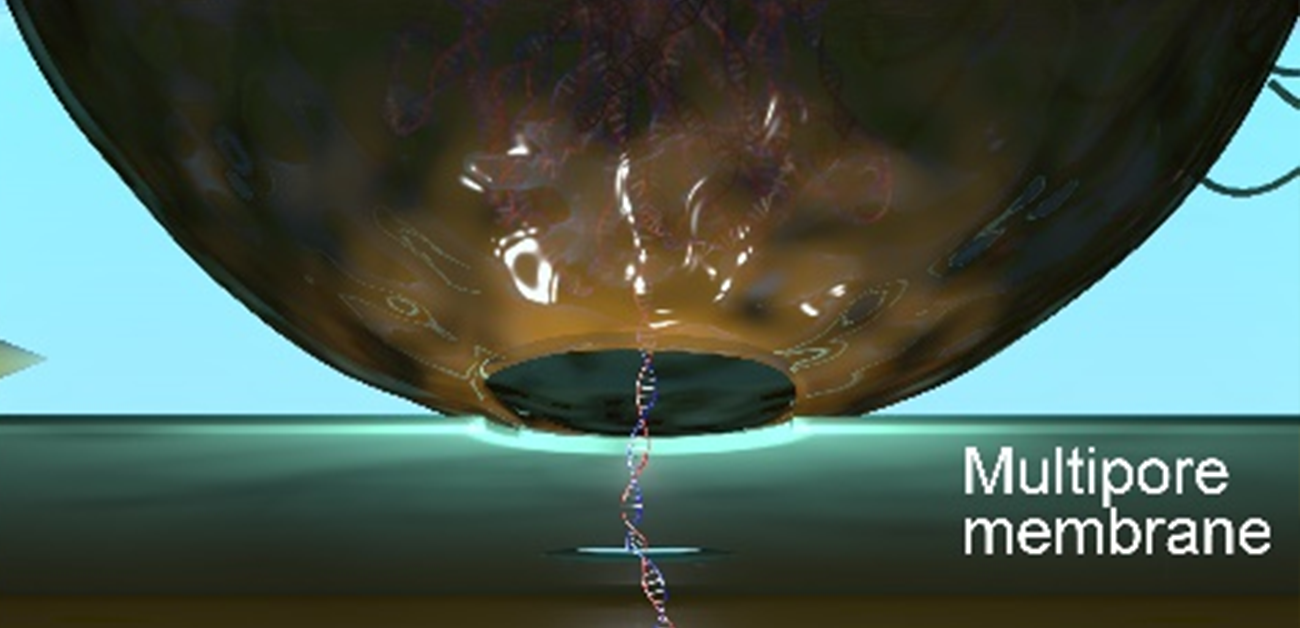
The researchers have created a 3D-integrated nanopore that can rupture cells immediately prior to measurement. The released molecules can be efficiently delivered to the sensing zone and measured without having to carry out any further steps that could introduce errors.
“Our sensor has two important parts. The first is a layer that contains numerous holes that are much smaller than a cell. An electrostatic field is used to rupture the cell and certain released substances can pass through holes while larger debris cannot, essentially providing a filter,” explains study first author Makusu Tsutsui. “Below this filter layer, separated by a spacer, is a single nanopore in a second membrane, where the measurements are made.”
When a voltage is applied, a current flows through the pore because of salt ions in the surrounding solution. This current is partially blocked when large DNA molecules are also passing through the pore, and the changes provide information about the large molecules. For example, whether the molecule—which can be millimeters in length—is folded.
“The filtering effect of our 3D-integrated nanopore prevents blockage of the measurement pore making it robust to use,” says study corresponding author Tomoji Kawai. “We therefore expect it to be used in new technologies for rapidly detecting mutant viruses at the genome level.”
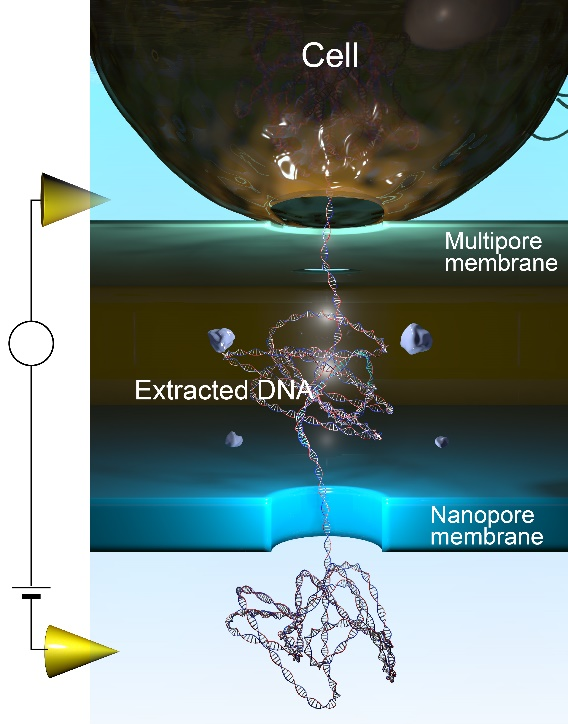
Fig. Extractions and detections of single-molecule DNA from a cell using a 3D-integrated nanopore. (credit: Makusu Tsutsui et al.)
The article, “Detecting single molecule deoxyribonucleic acid in a cell using a three-dimensionally integrated nanopore,” was published in Small Methods at DOI: https://doi.org/10.1002/smtd.202100542.
