Gene-network analysis is a valuable new tool for understanding Alzheimer’s disease
Researchers from Osaka University, Niigata University, and the National Center for Geriatrics and Gerontology propose a new method for identifying genes associated with neurodegeneration throughout the stages of Alzheimer’s disease
Alzheimer’s disease (AD) is known to involve interactions among many different genes, making it difficult to pinpoint the specific mechanisms. But now, researchers from Japan have found a new way to identify genes implicated in neurodegeneration in AD.
In a study published this month in Human Molecular Genetics , researchers from Osaka University, Niigata University, and the National Center for Geriatrics and Gerontology have revealed genes that trigger specific changes in connections between proteins, called protein domain networks (PDNs), that are significantly associated with neurodegeneration in AD.
Network analysis is effective for identifying features of AD in humans, as well as identifying genes that control pathological changes in network properties. The researchers conducted network analysis of changes in PDNs to determine whether this process could be used to reveal genetic changes associated with AD pathology throughout the different stages of the disease.
“PDNs can vary widely among tissues and cell types and have different structures in normal versus disease states,” explains Masataka Kikuchi, lead author of the study. “We wanted to see if we could use an integrated network approach to identify new genes associated with the collapse of PDNs in AD,” adds Michiko Sekiya, co-lead author.
To do this, the researchers examined protein interaction data and gene expression data from postmortem brain samples from patients with AD to generate PDNs. They found that the deterioration of PDNs occurred at specific stages of AD and identified RAC1 as a specific gene that plays a key role in the alteration of PDNs. They then examined the effects of deactivating RAC1 in the fruit fly Drosophila .
“We found reduced levels of RAC1 in postmortem brain samples from patients with AD, and these changes were validated by gene expression levels from a publicly available data set” says senior author Koichi M. Iijima. “Further, inhibiting RAC1 in Drosophila induced age-dependent changes in behavior, accompanied by neurodegeneration.”
In addition, they found that the number of interactions constituting PDNs in three regions from AD brains decreased with each stage of AD.
“These data provide further evidence that PDNs collapse during the progression of AD,” says Norikazu Hara, co-lead author. “Thus, changes in PDNs may be a key element of neuronal dysfunction and neurodegeneration that occurs in AD.”
These results indicate that an integrated network approach is advantageous in determining the factors that lead to neurodegeneration in AD, which may improve the chances of uncovering new biomarkers and therapeutic treatments.
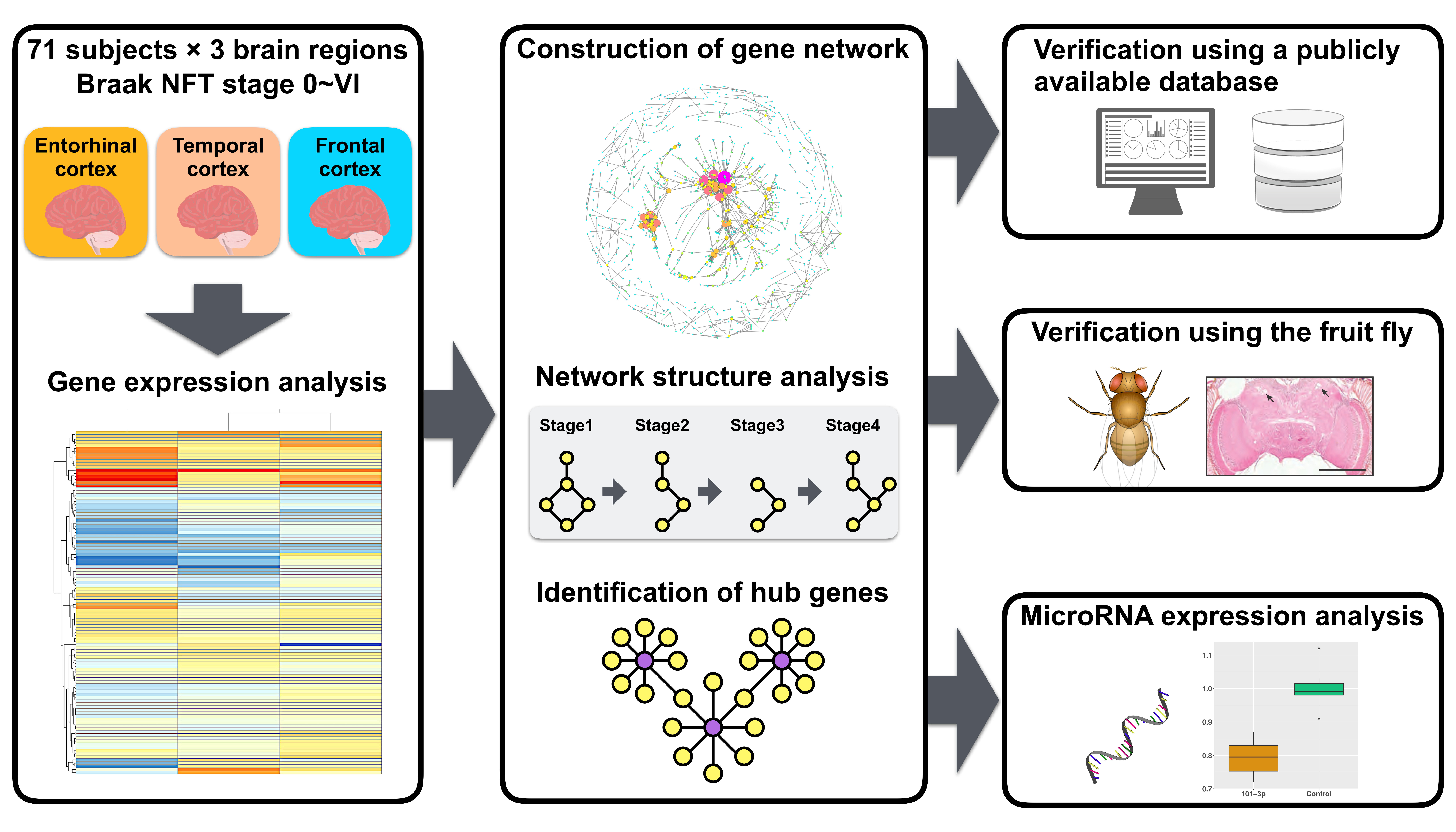
Fig. Overview of the present study.
The article, “Disruption of a RAC1-centred network is associated with Alzheimer’s disease pathology and causes age-dependent neurodegeneration” was published in Human Molecular Genetics at DOI: https://doi.org/10.1093/hmg/ddz320
