Tiny DNA Reader to Advance Development of Anticancer Drugs
Researchers from Osaka University use tiny probes and an electrical current to locate anticancer drug molecules incorporated into single strands of DNA
DNA is small. Really, really, small. So, when researchers want to study the structure of a single-stranded DNA, they can’t just pull out their microscopes: they have to get creative.
In a study published this week in Scientific Reports , researchers from Japan’s Osaka University explain how they came up with a really small solution to the challenge of studying anticancer drugs incorporated into single strands of DNA.
With almost half of us likely to develop cancer at some point in our lifetime, the need for novel and effective treatments has never been more critical. And while researchers are constantly developing new and improved therapies to kill cancer cells, or at least halt their replication, a limited understanding of precisely how these drugs work can sometimes make it difficult to advance otherwise promising treatments.
One such treatment, trifluridine, is an anticancer drug that gets incorporated into DNA as it replicates. While similar to thymine, one of the four nucleotides that make up DNA, trifluridine can’t bind to thymine’s partner nucleotide, adenine. This destabilizes the DNA molecule, resulting in aberrant gene expression and, ultimately, cell death.
But exactly where trifluridine gets incorporated into the DNA remains a mystery because it is not distinguished by traditional DNA sequencing methods, hampering efforts to fully understand and develop the technology.
Therefore, the team at Osaka University set about developing a DNA sequencing method that could distinguish the drug molecules from normal nucleotides in short strands of DNA. Using microscopic probes, the researchers passed an electrical current across a distance approximately 65,000 times smaller than a grain of sand—a gap just wide enough to fit a strand of DNA.
“Using this single-molecule quantum sequencing method, we successfully identified individual molecules in the DNA based on differences in electrical conductance,” explains lead author Takahito Ohshiro. “For the first time, we were able to directly detect anticancer drug molecules incorporated in the DNA.”
Importantly, the conductance of trifluridine was lower than that of the four native nucleotides, which also displayed divergent conductance values, allowing it to easily be distinguished in the DNA sequence. Based on these values, the researchers successfully sequenced single DNA strands of up to 21 nucleotides, pinpointing the exact insertion sites of trifluridine.
“Now that we have the ability to determine exactly where the drug is incorporated, we can develop a better understanding of the mechanism involved in DNA damage,” says senior author Masateru Taniguchi. “We expect that this technology will aid in the rapid development of new and more effective anticancer drugs.”
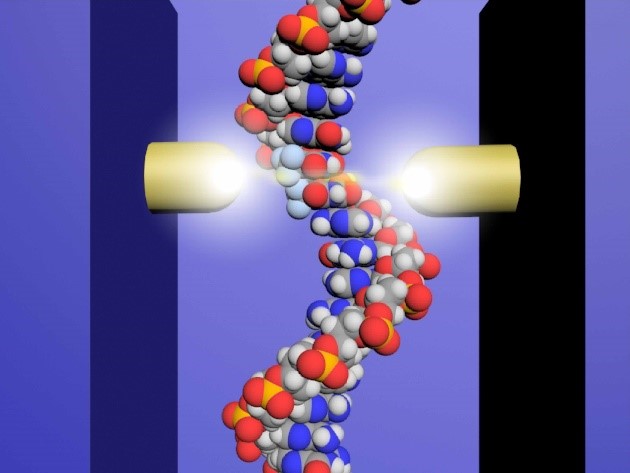
Figure. Diagram showing the operation of the single-molecule quantum sequencer.
(credit: Osaka University)
The article, “Direct Analysis of Incorporation of an Anticancer Drug into DNA at Single-Molecule Resolution,” was published in Scientific Reports at DOI: https://doi.org/10.1038/s41598-019-40504-x .
Related links
