
Artificial Intelligence Aids Automatic Monitoring of Single Molecules in Cells
Japanese researchers have developed an automated method of tracking single fluorescently labeled molecules in living cells, enabling large numbers of molecules to be analyzed and characterized rapidly and cost-effectively
To understand the mechanisms by which molecules act in cells, or the effects of drugs on them, it would be ideal to be able to track individual molecules, including where in the cell they are located and what modifications they undergo when conditions in the cell change. However, this has proven difficult with existing technologies, particularly given the amount of time required to perform such monitoring.
A research team centered at Osaka University, in collaboration with RIKEN, has developed a system that can overcome these difficulties by automatically searching for, focusing on, imaging, and tracking single molecules within living cells. The team showed that this approach could analyze hundreds of thousands of single molecules in hundreds of cells in a short period, providing reliable data on the status and dynamics of molecules of interest.
For the development of this method, reported in the journal Nature Communications , the team used an artificial intelligence-based system, involving the training of neural networks to learn to focus correctly on a sample and to automatically search for cells, followed by the tracking of single fluorescently labeled molecules with a total internal reflection fluorescence microscope.
The team tested this system on a receptor protein called EGFR, which is more or less free to move along the plasma membrane in which it is expressed depending on whether it has undergone a certain modification. Their findings showed that the system could differentiate between modifying and nonmodifying conditions by tracking the movements of single receptors in membranes.
“We used the results obtained by our system to calculate pharmacological parameters, such as those reflecting the efficacy of drugs and the speed with which molecules diffuse away from their initial location,” corresponding author Masashiro Ueda from Osaka University says. “The findings matched the values obtained in earlier studies using traditional labor-intensive methods, supporting the value of this system.”
“A major benefit of this approach is that the effects of ligands and inhibitors on a target can be quantified at the single-molecule level,” adds Ueda. “The automation provided by this approach means that a large number of targets exposed to such molecules can be characterized at low cost, increasing the reliability of the results.”
As future work, the team hopes to apply this system to the monitoring of single molecules elsewhere in the cell, such as in the nucleus and organelles, using other optical microscopes. The system should also be applicable clinically for reliable genome-wide screening and for pharmacological testing.
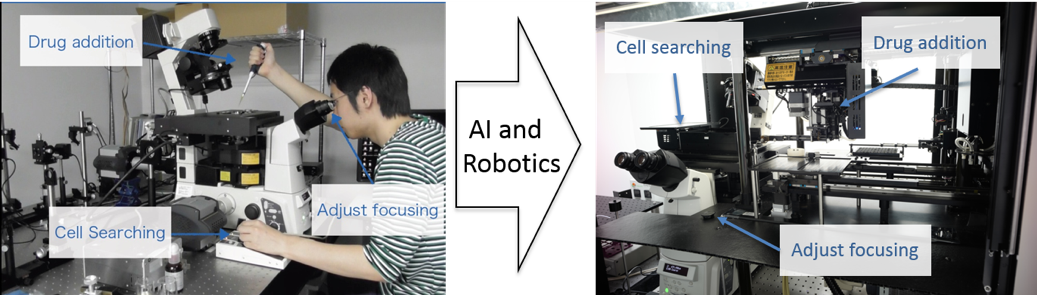
Fig. 1: Single-molecule imaging was fully automated in a manner that overcomes the need for technical expertise, such as precisely adjusting the focus and searching for analyzable cells, by an artificial intelligence-aided system.
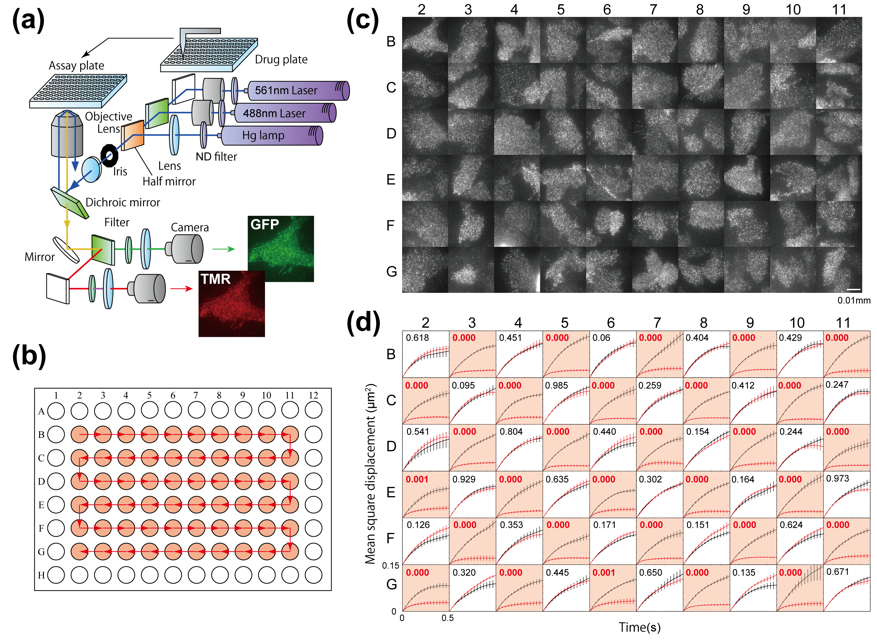
Fig. 2: The apparatus achieved large-scale single-molecule analysis on more than 1,600 cells per day, which had markedly higher efficiency than in previous techniques.
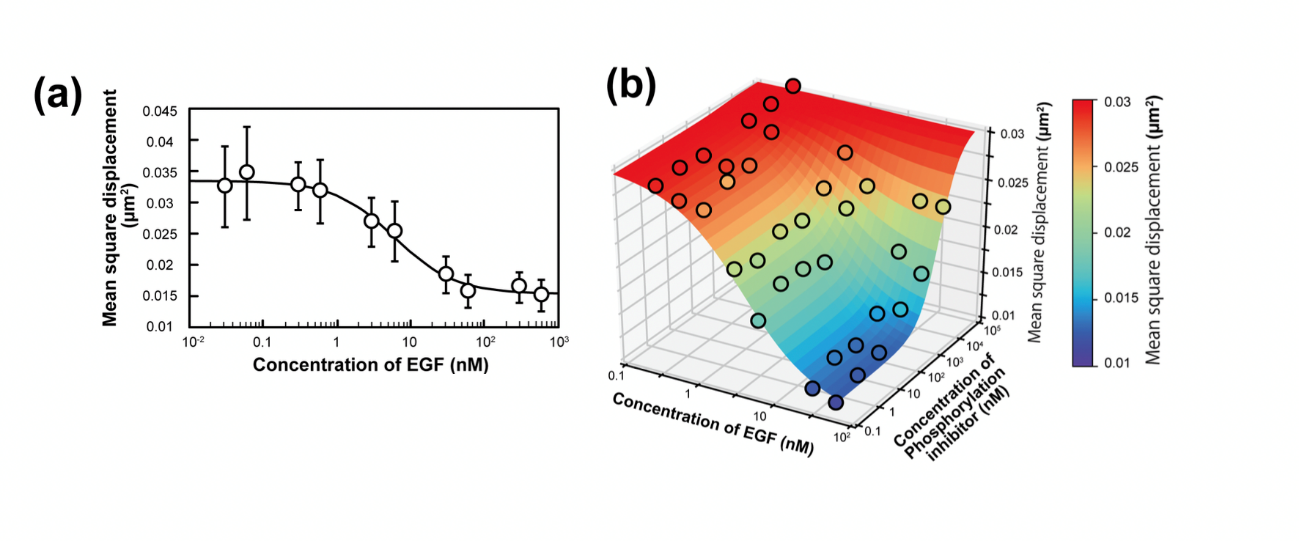
Fig. 3: The observed changes in the single-molecule behavior of epidermal growth factor receptor (EGFR) invoked by its ligand and kinase inhibitor were shown to be applicable to the measurement of pharmacological parameters.
The article “Automated single-molecule imaging in living cells” was published in Nature Communications, https://doi.org/10.1038/s41467-018-05524-7 .
Related links
