Excited motion of single protein molecule in aggregation process observed
Will lead to the development of new medicines for Alzheimer disease
A group of researchers led by Professor Yuji SASAKI at the Graduate School of Frontier Sciences, The University of Tokyo, together with researchers at Osaka University, Kobe University, and the Japan Synchrotron Radiation Research Institute, observed a random motion in and around a protein molecule in the aggregation processes of protein molecules (lysozyme) in a supersaturated solution by using a diffracted X-ray tracking (DXT) method.
This group examined these results and found that this dynamic motion generated an extremely small force field on the femto-Newton scale. This shows that there is rearrangement and relaxation of molecular networks accompanied by excited Brownian motion.
Abnormal protein aggregation, or amyloidosis, is known as the causative agent of some neurological disorders such as Alzheimer's disease, but their effective treatment methods have not been established. This may be because the behavior of protein molecules in solution in vivo is not fully understood.
This group focused on molecular assembly in supersaturated solutions with the goals of observing local molecular dynamic structures in protein solutions and establishing the measurement technology.
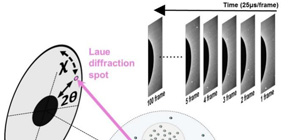
Previously this group used DXT as a method for high time resolution and high positional accuracy X-ray single molecule observation. This time, it was found that DXT could be used for dynamic measurement of nanoscale microscopic structures in protein molecules.
This group performed DXT measurements in a supersaturated solution, a precursor state to crystal formation, at High Flux Beamline (BL40XU) of the large-scale synchrotron radiation facility SPring-8. As a result, they measured rotational dynamic structures of a single gold nanocrystal particle labeled to a specific site in individual protein molecules with high accuracy and high speed.
These results suggest the existence of ion networks in a supersaturated solution. Since the group led by Prof. Sasaki confirmed Ion networks for inorganic supersaturated solutions in 2015, in this study, the dynamic structure of ion networks common to organic, inorganic, and proteins was confirmed. It was also confirmed that an extremely small force field on the femto-Newton scale was involved in Brownian motion.
This group has made it possible to observe single molecules involved in protein aggregation, which is related to the development of Alzheimer disease. The group’s achievement will lead to the development of new treatment strategy using supersaturated conditions in the future.
Abstract
Proteins in solution are conventionally considered macromolecules. Dynamic microscopic structures in supersaturated protein solutions have received increasing attention in the study of protein crystallisation and the formation of misfolded aggregates. Here, we present a method for observing rotational dynamic structures that can detect the interaction of nanoscale lysozyme protein networks via diffracted X-ray tracking (DXT). Our DXT analysis demonstrated that the rearrangement behaviours of lysozyme networks or clusters, which are driven by local density and concentration fluctuations, generate force fields on the femtonewton to attonewton (fN – aN) scale. This quantitative parameter was previously observed in our experiments on supersaturated inorganic solutions. This commonality provides a way to clarify the solution structures of a variety of supersaturated solutions as well as to control nucleation and crystallisation in supersaturated solutions.
Figure 1
To learn more about this research, please view the full research report entitled " Nanoscale Dynamics of Protein Assembly Networks in Supersaturated Solutions " at this page of the Scientific Reports website.
Related link
