Mechanisms of threshold or switch-like gene expression response clarified by omics data analysis and mathematical modeling
A group of researchers from the Institute for Protein Research of Osaka University, the RIKEN Center for Integrative Medical Sciences, the RIKEN Center for Biosystems Dynamics Research, the Medical Research Institute of Tokyo Medical and Dental University, and The University of Tokyo has clarified the molecular mechanisms of a rapid antigen response of B cells in the immune system through omics data analysis and mathematical modeling.
B cells not only produce antibodies but also actively regulate immune responses by producing cytokines, which play a crucial role in the regulation of immune responses, such as the amount and timing of gene expression, but how gene expression is controlled was not well understood.
This research group stimulated primary mouse B cells for 1 h with anti-immunoglobulin M (IgM) and performed multi-omics analysis of gene expression and epigenetic modifications obtained from cells.
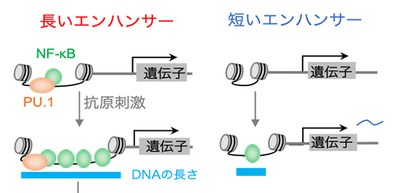
In stimulated B cells, a greater number of NF-κB (Nuclear factor kappa-light-chain-enhancer of activated B cells) molecules bound to enhancers (regulatory DNA sequences that provide binding sites for proteins that help activate transcription) than in unstimulated B cells. NF-κB, a transcription factor (TF), activated super enhancers (SEs) and typical enhancers (TEs) and triggered threshold and graded gene expression, respectively. SEs with longer DNA have larger capacity for multiple NF-κB binding.
Their analysis indicated that the increased NF-κB binding to SEs associated with a higher fold change (“fold change” is an important quantity to show the change of gene expression levels) resulted from longer DNA.
Their mathematical modeling revealed that SE-associated genes triggered threshold or switch-like gene expression. The number of NF-κB molecules bound to DNA (n=12) associated with the SE region of Irf4, an NF-κB target gene that promotes the activation of B cells in their proliferation and differentiation. NF-κB target gene Irf4 showed a larger fold change in response to the increased dosage of anti-IgM.
This group also found that the NF-κB-PU.1 complex acted as a seed in the enhancer regions with enriched NF-κB binding and triggered sequential NF-κB binding. The higher fold change results from longer DNA, whereas the threshold response is predefined by the coexistence of PU.1, a transcription factor, and NF-κB in a SE before cell stimulation.
Since NF-κB is translocated from the cytoplasm into the nucleus upon cell stimulation via the NF-κB signaling pathway, which is activated by B cell receptors (BCRs), this shows that the NF-κB signaling pathway to control NF-κB in the nucleus also plays an important role in antigen responses.
The NF-κB SE-mediated gene expression resulted in a larger distribution of transcript levels in a cell population than that mediated by TE. This suggests an additional role of SEs for divergent gene expression and their research results indicate that the cell differentially uses SEs and TEs strategically to adapt to new environmental changes.
NF-κB is not only required for maintaining normal immune systems but also is involved in the growth of some types of B-cell lymphomas and cancers with a poor prognosis. The omics data analysis and numerical modeling used in this study can be applied to the clarification of molecular mechanisms of various diseases, identification of marker genes, and drug development.
Figure 1
Figure 2
The article, “The Number of Transcription Factors at an Enhancer Determines Switch-like Gene Expression,” was published in Cell Reports at DOI: https://doi.org/10.1016/j.celrep.2020.107724.
