In vivo genome editing technique for directly treating intractable genetic diseases in tissues
A group of researchers from Osaka University and the Salk Institute for Biological Studies (U.S.) developed an in vivo knock-in methodology, Single homology Arm donor mediated intron-Targeting Integration (SATI), using a gene-editing tool CRISPRCas9.
Pathogenic gene mutations cause inherited diseases, most of which are intractable. Thus, genome technology to design and modify genome sequences by innate double-strand break (DSB) repair machineries drew attention and programmable site-specific nucleases such as CRISPR-Cas9 were developed for use of genome editing.
There are two primary mechanisms to repair DSBs: (a) homologous recombination (HR) or homology-directed repair (HDR), which causes recombination using a homologous sequence as a template for repair DNA synthesis and (b) non-homologous end joining (NHEJ), which legates broken ends of DSBs without the need for a homologous template.
Genomic engineering by CRISPR-Cas9 allows researchers to cleave DSBs at the on-target sites. Using CRISPR-Cas9, gene knock-out and knock-in can be achieved after introducing DSBs at the target site. However, the technology was incapable of targeted knock-in in non-dividing cells, such as cells of the eye, brain, pancreas, or heart.
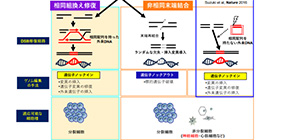
To overcome the limitations of HDR-mediated genome editing, in 2016, using the NHEJ pathway, this group developed another genome editing technology CRISPR/Cas9-based homology-independent targeted integration (HITI), which achieved high efficiency that was not achieved through the HDR pathway.
In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration (https://www.nature.com/articles/nature20565)
HITI allowed for efficient targeted knock-ins in both dividing and non-dividing cells in vitro and were effective for targeting loss-of-function mutations caused by large deletions, but couldn’t repair point or frameshift mutations.
In this study, the group improved HITI, developing a method called Single homology Arm donor mediated intron-Targeting Integration (SATI), which allows for the targeting of a broad range of mutations and cell types. SATI-mediated knock-in has been achieved in both dividing and non-dividing cells, as is the case with HITI. However, one-armed HDR (oaHDR)-mediated integration was observed in some cells. Thus, by creating a unique vector structure, they developed SATI that allowed for repair via either HITI- or oaHDR-mediated targeted gene knock-in.
In order to confirm the effectiveness of SATI for systemic genetic diseases, the group systemically delivered Adeno-associated virus (AAV)-Progeria-SATI, together with an AAV expressing Cas9, via intravenous (IV) injection into neonatal progeria mice. They observed gene repair in the kidney and heart, confirming meliorated aging-associated phenotypes and extended animal lifespan. That is, they succeeded in developing a systematic gene therapy for genetic mutations, including the in vivo correction of a dominant point mutation, which was difficult to repair using existing in vivo genome-editing tools.
Further improvement of this technique will lead to the development of medical care to directly repair mutations that cause refractory inherited diseases, which have anomalies in the adult nervous system, heart, muscles, retina, or whole body.
Figure 1
Figure 2
Figure 3
Figure 4
The article “Precise in vivo genome editing via single homology arm donor mediated intron-targeting gene integration for genetic disease correction” was published in Cell Research at DOI: https://doi.org/10.1038/s41422-019-0213-0.
Related links
