Structural fluctuations in proteins determine which direction bacteria are moving
A step forward to artificial nanomachines capable of controlling travelling direction
A group of researchers from Nagoya University, Osaka University, and Nagahama Institute of Bio-Science and Technology clarified the structural dynamics of FliG, one of the proteins that make up the flagellar motor that a bacterium uses for movement, played a critical role in determining a rotational direction of the motor.
Bacteria swim in the water by rotating their helical flagellar filament extending from the cell’s surface like a screw for propulsion. They move by rotating flagellar filaments in either a counterclockwise (CCW) or clockwise (CW) direction. If all the motors rotate CCW, the filaments drive the cell forward and if all the motors rotate CW, the filaments drive the cell backward.
The drive part of a flagellar motor consists of a rotor and a stator and torque is generated by the interaction of stator units with the rotor. Signals from receptors on the cell membrane are transmitted to chemotaxis signal protein CheY, which binds to the rotor, changing the rotational direction of the motor.
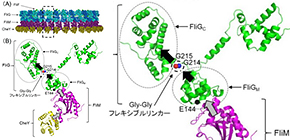
FliG is one of three proteins (FliG, FliM, FliN) that form the rotor-mounted switch complex (C ring). The researchers demonstrated that the structure of FliG in the vicinity of the EHPQR motif, the FliM binding site in FliGM, differed between the CCW and CW direction of the flagellar filaments. Of the proteins that make up a rotor, FliG, FliM, and FliN interact to form the C ring, and the ring controls the switching mechanism for regulating the generation and direction of flagellar rotation.
In this study, the researchers introduced multiple types of mutants such as E144D that produce abnormalities in rotational direction control to FliG of marine Vibrio to examine the relationship between structural changes in FliG and rotational direction control. When they introduced the mutants to the hinge linker region between FliGM and FliGC, CW- or CCW-locked rotation or stoppage after the repetition of movement in both forward and backward directions were observed in bacteria.
In the linker region, the conformational change in mutant FliGMC was more restricted than that in wild-type FliGMC. Analysis of mutations suggested that the flexible linker played a decisive role in determining the rotational direction of the flagellar motor. Since abnormalities were observed in signal response of the E144D mutant, the researchers speculated that mutations affected the interaction with FliM rather than the conformational change of FliG.
The group clarified that the structure of FliG and the interaction between FliM and FliG played an important role in determining flagellar rotational direction and switching. If this group’s achievements lead to the clarification of a mechanism behind the flagellar rotational direction, it will become possible to design artificial nanomachines capable of controlling their direction at will and apply these results to various fields.
Abstract
FliG, which is composed of three distinctive domains, N-terminal (N), middle (M), and C-terminal (C), is an essential rotor component that generates torque and determines rotational direction. To determine the role of FliG in determining flagellar rotational direction, we prepared rotational biased mutants of fliG in Vibrio alginolyticus. The E144D mutant, whose residue is belonging to the EHPQR-motif in FliG M , exhibited an increased number of switching events. This phenotype generated a response similar to the phenol-repellent response in chemotaxis. To clarify the effect of E144D mutation on the rotational switching, we combined the mutation with other che mutations (G214S, G215A and A282T) in FliG. Two of the double mutants suppressed the rotational biased phenotype. To gain structural insight into the mutations, we performed molecular dynamic simulations of the FliG MC domain, based on the crystal structure of Thermotoga maritima FliG and nuclear magnetic resonance analysis. Furthermore, we examined the swimming behavior of the fliG mutants lacking CheY. The results suggested that the conformation of FliG in E144D mutant was similar to that in the wild type. However, that of G214S and G215A caused a steric hindrance in FliG. The conformational change in FliG M triggered by binding CheY may lead to a rapid change of direction and may occur in both directional states.
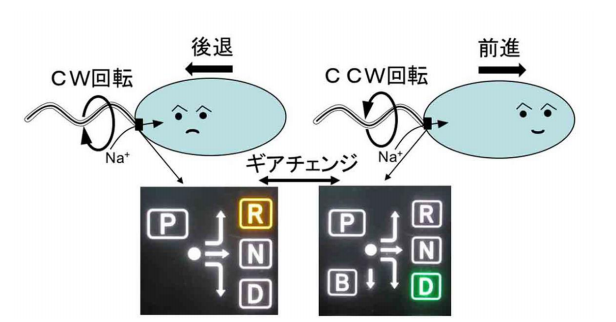
Figure 1
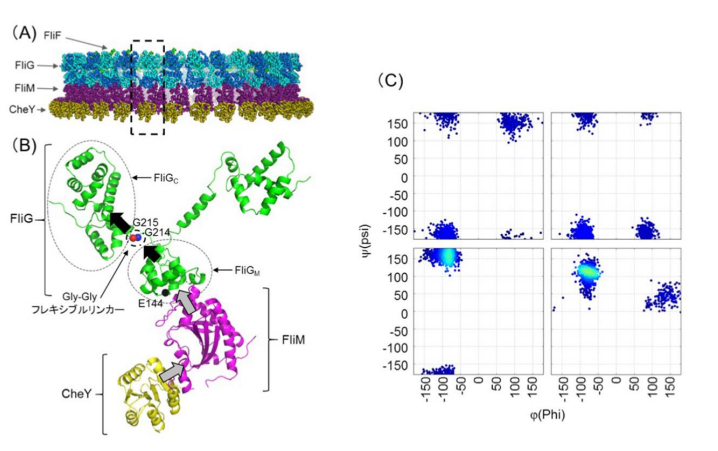
Figure 2
The article "Rotational direction of flagellar motor from the conformation of FliGmiddle domain in marine Vibrio” was published in Scientific Reports at DOI: https://doi.org/10.1038/s41598-018-35902-6 .
Related links
