Computer Simulation of Protein Synthesis Reveals Awesome Complexity of Cell Machinery
Osaka University-led researchers model protein production by system of 241 components based on E. coli bacteria
Life depends on proteins. These molecules are produced continually in our cells, which act as microscopic production lines – but the process is so complex we have barely begun to understand it. Exploring protein synthesis may, however, be the key to revealing how the body controls the thousands of reactions occurring simultaneously inside us.
Now, a Japanese research team led by Osaka University and RIKEN has run a computer simulation of a small protein (peptide) being produced by bacterial translation machinery, confirming a “molecular toolkit” as highly sophisticated even for simple lifeforms. A report of their findings was recently published in Proc Natl Acad Sci U S A .
Full-size proteins are too large to model in detail. However, a tripeptide (made from three amino acids: “Met-Gly-Gly”) can be produced by a set of 27 molecules in the bacteria E. coli. The researchers simulated this process, also known as an in-vitro translation (IVT), by modeling the chemical reactions on a computer.
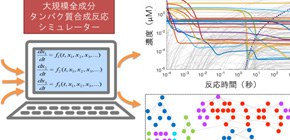
Even for this simple tripeptide, over 900 reactions were needed, with over 200 chemicals formed as by-products. These reactions occurred at wildly different speeds, and the slowest acted as bottlenecks on the overall process. The researchers found some reaction rates from earlier experiments, while others had to be assumed. Then, they simulated 1,000 seconds of the resulting chemical dance.
“We were surprised that even with so many assumptions and simplifications, the simulation accurately described the kinetics of Met-Gly-Gly production,” study lead author Tomoaki Matsuura says. “When we increased the amounts of some initial components, the rate of synthesis increased, in line with real IVT.”
By tracking the amount of each chemical present, the researchers found that many chemicals reached a stable level (termed steady-state), however, only to suddenly decrease again later on. In fact, this collapse happened twice – after 0.5 sec and again after 10 sec.
The researchers believe this non-linear behavior, wherein chemicals reach a “stable state” in the system, but are later recruited back to “active state”, is typical of biochemistry.
“The peak, collapse, and further peaking of steady states is likely to occur in other processes, such as glucose metabolism,” Matsuura says.
Matsuura adds that the simulations could be extended to different proteins, including larger ones. “By comparing our simulations with IVT experiments,” he says, “we can improve the IVT systems – which in turn will help us improve the simulations. The awesome complexity of this ‘simple’ synthesis reminds us that we are just beginning to understand life at the molecular level.”
Abstract
To elucidate the dynamic features of a biologically relevant large-scale reaction network, we constructed a computational model of minimal protein synthesis consisting of 241 components and 968 reactions that synthesize the Met-Gly-Gly (MGG) peptide based on an Escherichia coli-based reconstituted in vitro protein synthesis system. We performed a simulation using parameters collected primarily from the literature and found that the rate of MGG peptide synthesis becomes nearly constant in minutes, thus achieving a steady state similar to experimental observations. In addition, concentration changes to 70% of the components, including intermediates, reached a plateau in a few minutes. However, the concentration change of each component exhibits several temporal plateaus, or a quasi-stationary state (QSS), before reaching the final plateau. To understand these complex dynamics, we focused on whether the components reached a QSS, mapped the arrangement of components in a QSS in the entire reaction network structure, and investigated time-dependent changes. We found that components in a QSS form clusters that grow over time but not in a linear fashion, and that this process involves the collapse and regrowth of clusters before the formation of a final large single cluster. These observations might commonly occur in other large-scale biological reaction networks. This developed analysis might be useful for understanding large-scale biological reactions by visualizing complex dynamics, thereby extracting the characteristics of the reaction network, including phase transitions.

Figure 1. Developed a large-scale whole component protein synthesis translation simulator

Figure 2. A result of protein synthesis simulation. Concentration changes of 241 components are shown.

Figure 3. Growth and collapse of the network composed of QSS components. Circle indicates the QSS component and line indicates the connection between the components.
To learn more about this research, please view the full research report entitled “ Reaction dynamics analysis of a reconstituted Escherichia coli protein translation system by computational modeling ” at this page of the Proceedings of the National Academy of Sciences of the United States of America website.
Related link
