Successful recording of 3D movie of atom movement in proteins at an x-ray free electron laser (XFEL)
Mechanism behind transport of hydrogen ions following photoactivation clarified
A group of researchers from Kyoto University, RIKEN, Japan Synchrotron Radiation Research Institute, The University of Tokyo, Kobe University, Osaka University, and the University of Gothenburg, succeeded in filming movement of membrane proteins frame by frame at an atomic level by using high-quality light at the RIKEN’s X-ray Free Electron Laser (XFEL), SPring-8 Angstrom Compact Free Electron Laser (SACLA), a world first.
Proteins play an important role in life and change their shape when functioning. However, with conventional X-ray diffraction methods, only a resting state could be observed and live imaging was rarely feasible. This group developed an experiment device capable of examining shapes of moving proteins by using XFEL facilities at SACLA. This device is the combination of serial femtosecond crystallography (SFX) and pump-probe spectroscopy.
Furthermore, their experimental device visualized membrane protein movement by using light-driven membrane protein bacteriorhodopsin, which transports hydrogen ions after photoactivation. This group succeeded in filming conformational changes of membrane proteins at many time points on the nano- to millisecond time scale, clarifying how protein structures and water molecules transport hydrogen ions. This group’s achievement has made it possible to clarify a moving state of proteins at an atomic level. This will lead to a wide range of applications in medical care and engineering, such as design and development of medicines and functional molecules.
Abstract
Bacteriorhodopsin (bR) is a light-driven proton pump and a model membrane transport protein. We used time-resolved serial femtosecond crystallography at an x-ray free electron laser to visualize conformational changes in bR from nanoseconds to milliseconds following photoactivation. An initially twisted retinal chromophore displaces a conserved tryptophan residue of transmembrane helix F on the cytoplasmic side of the protein while dislodging a key water molecule on the extracellular side. The resulting cascade of structural changes throughout the protein shows how motions are choreographed as bR transports protons uphill against a transmembrane concentration gradient.
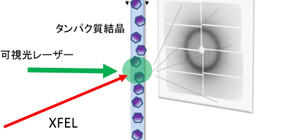
Fig.1. Schematic drawing of experimental method to capture structural changes of proteins. The microcrystals are continuously fed to the XFEL irradiation region and irradiated with a visible light laser to start the protein reaction. Changes in proteins occurring after nanoseconds to milliseconds after the visible light irradiation can be examined by obtaining diffraction images by XFEL.
Fig.2. Time-dependent structural change of bacteriorhodopsin protein. Structural changes of the protein were tracked at 13 time points in nanoseconds to milliseconds after the protein began to react with light. Analysis of the obtained structure revealed that it mainly takes four kinds of intermediate structures. The portion drawn with a purple ribbon shows the main chain of the protein. The blue and yellow parts inside the structure show that changes have occurred compared to the structure before movement and that the atoms in the yellow part have moved to the blue part.
Fig.3. Structural change around retinal: before visible light irradiation (pink) and 36.2 microseconds after irradiation (orange). The movements of retinal, protein and water molecules (indicated by W) are shown.
To learn more about this research, please view the full research report entitled “ A Three Dimensional Movie of Structural Changes in Bacteriorhodopsin ” at this page of the Science website.
Related link
