Researchers discover key mechanism of DNA replication
Researchers from Osaka University in Japan have uncovered a key control mechanism of DNA replication with potential implications for better understanding how cells maintain genetic information to prevent diseases or cancer.
The team published their results in July in The EMBO Journal , the flagship publication of the European Molecular Biology Organization.
“DNA replication initiates from a number of different locations known as replication origins,” said author Hisao Masukata, a professor of biology in the Graduate School of Science at Osaka University.
To ensure faithful duplication of the cell’s genetic blueprints, replication begins at distinctly different times as well as locations. Each cell’s genetic material is arranged into specific “files” called chromosomes. The “tabs” labeling these files are called telomeres—they help ensure the DNA sequences begin and end in the correct order and at the correct times.
The mechanism underlying this spatiotemporal control was unknown, but the team suspected an answer could be found by more closely examining the telomeres.
“Relation between time and space of chromosomal DNA replication was a long-term question and there were controversial observations,” Masukata said. “The main problem we set out to solve was whether a specific intra-nuclear compartment plays a role in regulation of DNA replication.”
The researchers found that telomeres don’t work on their own. Proteins bind to them, attaching messages of timing and other work notices. One protein, called Taz1, is of particular importance: It binds to the telomeres and a small number of Taz1 binds to the internal regions of chromosomes. The researchers were able to observe the internal regions and the telomeres by using different fluorescent proteins that light up in different colors. They used a microscope to follow their movement as the internal regions associate with telomeres during the replication process in live cells.
They saw that Taz1, together with its partners draws the internal regions to telomeres, like a paper clip to prevent free movement of papers.
Masukata calls this clip “tethering.” It links a replication timing program of the internal regions to that of later replicating telomeres. However, he noted, the replication origins—the location of replication initiations—are only tethered to telomeres in the beginning phases of the cell cycle. It’s still unclear how the tethering is regulated by the cell cycle.
By better understanding this intricate process of cell duplication, including when and how specific genetic information is copied, scientists may be able to inform future research into how cells maintain genetic information to prevent diseases or cancer.
“Our ultimate goal is to understand the principle and the significance of [the organization of DNA replication]. Our results emphasized the importance of telomeres for structures and functions of whole chromosome regions,” Masukata said. “Because telomeres are essential and highly conserved among [most organisms], they might play many important roles in various levels of chromosome architecture and function.”

Fig. 1, Model of intra-nuclear localization of replication origins in fission yeast nucleus.

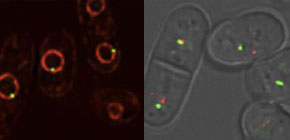
Fig. 2, Late-replication origins (green) are localized at the nuclear membrane (red).
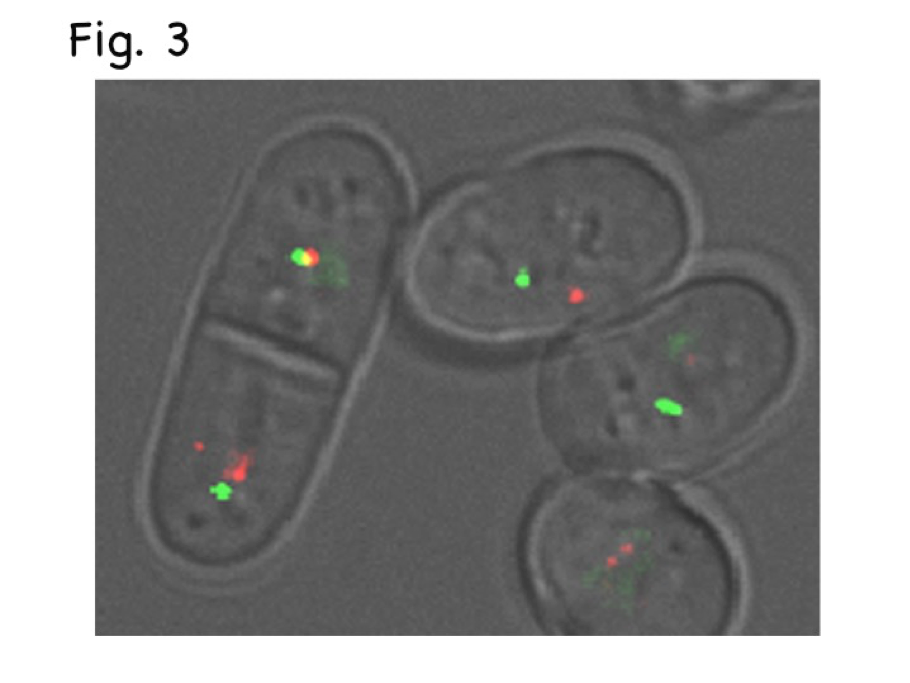
Fig. 3, Late-replication origin (green) and a telomere (red) partially overlap (creating yellow) in the left G1/S cell, while origins and telomeres are separate in right G2 cells.
The article, “Shelterin promotes tethering of late replication origins to telomeres for replication-timing control” was published in EMBO Journal , https://dx.doi.org/10.15252/embj.201898997 .
Related links
