A quick and easy detection of drug-resistant bacteria in clinical practice is available at a low cost
A new method for preventing the spread of such bacteria
A group of researchers led by Lecturer AKEDA Yukihiro and Professor TOMONO Kazunori at the Osaka University Hospital, in cooperation with the Research Institute for Microbial Diseases, Osaka University, developed a quick and easy method for detecting Carbapenem-resistant enterobacteriaceae (CRE), a type of bacteria that are highly resistant to many antibiotics. The spread of CRE in healthcare settings is a serious medical problem. This method simultaneously detects multiple genes involved in Carbapenem resistance using a technology Printed Array Strip (PAS) based on the Single-stranded Tag Hybridization (STH) method.
Prevention of the spread of drug-resistant bacteria is being promoted all over the world. CRE, in particular, are considered an urgent public health threat. CRE are bacteria that are resistant to Carbapenem-resistant antibiotics, which are considered to be the drugs of last resort for bacterial infectious diseases. CRE are resistant not only to Carbapenem, but also to almost all antimicrobial drugs, so the condition of CRE-infected patients can likely become severe. The fatality rate in case of bloodstream infection reaches 50%.
Carbapenemase genes related to Carbapenem resistance are carried not on bacteria’s chromosomal DNA, but on plasmids, so they can make other enterobacteriaceae without antibiotic resistance become Carbapenem-resistant. Thus, preventing CRE proliferation is an issue of utmost importance.
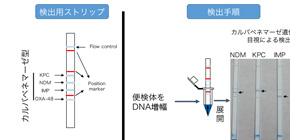
With this method, it’s possible to detect multiple Carbapenemase genes in CRE by visual observation within 15 minutes after DNA amplification of drug-resistant bacteria using stool specimens. As it’s also possible to correctly determine the presence of Carbapenemase genes even if multiple specimens are mixed, researchers can determine whether CRE is present or not in patients at risk for being exposed to CRE at a low cost.
For conventional CRE detection, expensive equipment and long response time were needed, so it was hard to detect CRE in clinical practice. However, this method will allow easy implementation of CRE infection control even in not well-equipped places, such as developing countries.
Abstract
A PCR-dipstick chromatography technique was designed and evaluated for differential identification of bla NDM , bla KPC , bla IMP , and bla OXA-48 carbapenemase genes directly in stool specimens within 2 h. It is a DNA-DNA hybridization-based detection system where PCR products can be easily interpreted by visual observation without electrophoresis. The PCR-dipstick showed high sensitivity (93.3%) and specificity (99.1%) in directly detecting carbapenemase genes in stool specimens compared with multiplex PCR for genomic DNA of the isolates from those stool specimens.
Figure 1
To learn more about this research, please view the full research report entitled " PCR-Dipstick Chromatography for Differential Detection of Carbapenemase Genes Directly in Stool Specimens " at this page of the Antimicrobial Agents and Chemotherapy website.
Related link
- Infection Control Team, Osaka University Hospital (link in Japanese)
