
New Integrated Analytical Approach Reveals Molecules Involved in Disease
Researchers at Osaka University and colleagues establish a method that integrates genetic and molecular data to reveal tissue-specific features that have novel associations with diseases
Technological advances have enabled scientists to obtain massive amounts of data on different information-carrying molecules in cells and tissues, such as DNA, proteins and various forms of RNA. However, to date, it has been difficult to perform integrated analyses of such information to further our knowledge on the molecules and processes involved in the development of particular diseases.
Researchers from an Osaka University-led collaboration have made a major contribution to our understanding of the complex relationships among the components of cells and tissues. They have established a new analytical approach called MIGWAS that integrates information from two different sources. One of these is associations between genetic variants and diseases and other traits, while the other is data about networks formed between small regulatory molecules called miRNAs and the genes that they target. The team showed that this in silico screening approach provides improved power to reveal novel cellular components associated with diseases and other traits, tissue-specifically.
The use of miRNA data in this way was prompted by accumulating findings on the importance of miRNAs in various physiological functions and diseases. miRNAs are small products of DNA that do not themselves encode proteins, but instead bind to mRNAs to regulate gene expression. In this latest study, the researchers employed previously obtained data on the expression of around 1,800 miRNAs in different cells. They searched for overlaps between these miRNAs and the genes they target on the one hand, and genes identified elsewhere as being linked to diseases and other traits by studies focusing directly on the genetic code on the other.
“It has been difficult to obtain convincing data about the effects of miRNAs as their short length means that they are encoded by only a very small part of the genome,” lead author Saori Sakaue says. “Our MIGWAS approach linking findings from genome-wide association studies and miRNA-target networks helps to overcome this. It successfully obtained significant results for miRNA/target gene enrichment for traits such as height and type 2 diabetes across different ethnic groups.”
The team attempted to validate this approach by focusing on the disease rheumatoid arthritis (RA), using data on single-nucleotide polymorphisms from nearly 20,000 RA cases and over 60,000 corresponding controls. This revealed four miRNAs linked to RA that were also identified by the MIGWAS pipeline, one of which was significantly highly expressed in RA patients.
“The analytical power of our approach shows its potential for finding new networks of miRNAs and their target genes linked to traits and diseases, in a tissue-specific way,” corresponding author Yukinori Okada says. “This should make it easier to find new targets that we can focus on for developing therapeutic strategies for a wide range of diseases.”
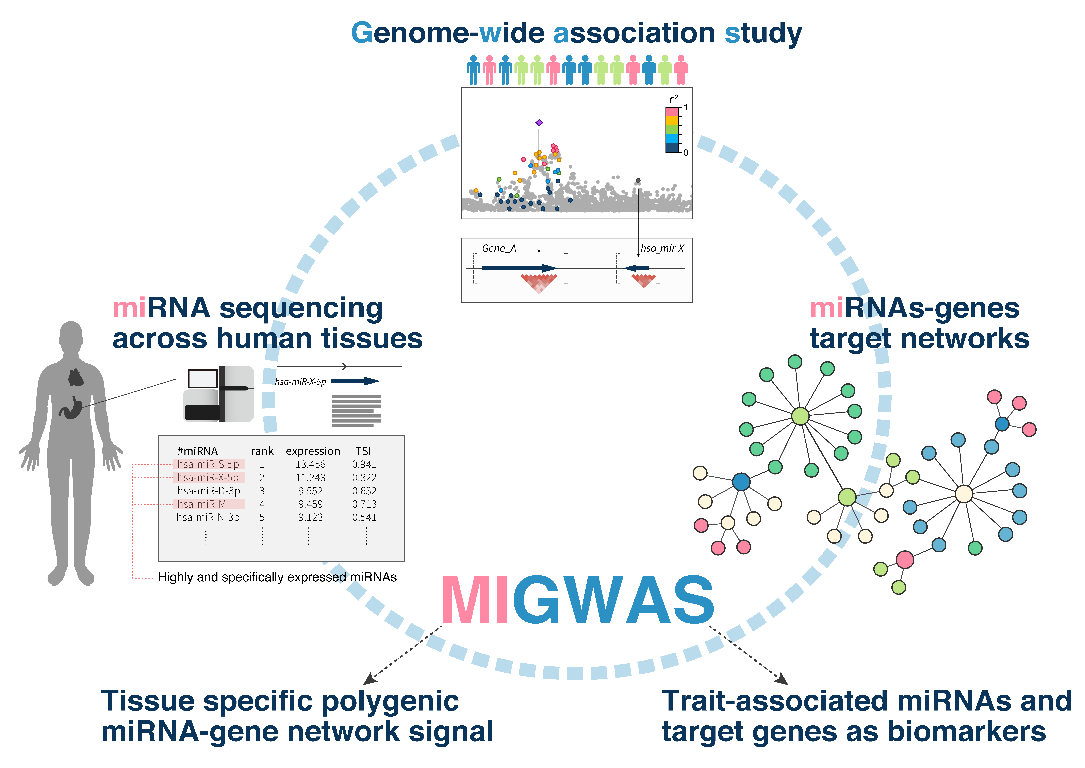
Figure: The MIGWAS pipeline integrates GWAS results, cell-type specific expression profiles of miRNAs, and in silico miRNA-gene target prediction algorithms to identify tissue specific contribution of miRNA to diseases and to detect biomarker miRNAs.
The article “Integration of genetics and miRNA-target gene network identified disease biology implicated in tissue specificity” is published in Nucleic Acids Research at DOI: https://doi.org/10.1093/nar/gky1066 .
Related links
