Elucidating unknown mechanisms behind protein function
Mechanisms of auto-inhibition of gene expression visualized
A group of researchers led by Assistant Professor KASAHARA Kota at Ritsumeikan University, Professor HIGO Junichi and Professor NAKAMURA Haruki at Osaka University, and Assistant Professor SHIINA Masaaki and Professor OGATA Kazuhiro at Yokohama City University, clarified that protein function was regulated by post-translational modifications (PTMs) (i.e., phosphorylation) of proteins with intrinsically disordered regions (IDRs) by using molecular simulations and biochemical experiments with mutational studies.
Humans are composed of various different types of body tissues and each of these tissues are composed of functionally differentiated cells. Each cell selectively reads information about proteins necessary for its function and synthesizes them. Proteins known as transcription factors bind to DNA to control the rate of transcription of genetic information from DNA.
This group focused on transcription factor Ets1, which regulates protein synthesis that is important for functions of blood cells. It is reported that abnormalities in the Ets1 protein are related to tumor invasion and metastasis.
It is thought that the functionalization of a protein of interest is required for the formation of its specific structure; however, IDRs that do not assume unique three-dimensional structures by themselves are abundantly found in proteins of higher organisms including humans. In IDRs, after chemical modification of proteins (post-translational modification (PTM)), protein functions are regulated as if the chemical modifications determine which genes are active and which are turned off.
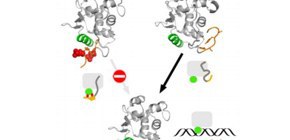
Ets1 also has IDRs and, once they are phosphorylated, it stops binding to DNA (turned off). However, the specific structures of IDRs that turn genes on had not been detected in conventional experiments and their behavior had been unknown.
This group clarified that IDRs did not change their forms freely, but rather turned genes on by forming specific structures depending on the phosphorylation of IDRs.
IDRs of Ets1 play their biological roles by forming different structures according the phosphorylation state of each Ets1 fragment. The regions with structures that were unidentified in experiments have not been a target for drug discovery, but this group’s achievements have enhanced the prospect that IDRs, even if their structures are not determined, which could become a target for drug discovery. The researchers will also study IDRs of other proteins which are involved in the onset and development of diseases and clarify their molecular structures in terms of their application to drug discovery.
Abstract
Multi-modal interactions are frequently observed in intrinsically disordered regions (IDRs) of proteins upon binding to their partners. In many cases, post-translational modifications in IDRs are accompanied by coupled folding and binding. From both molecular simulations and biochemical experiments with mutational studies, we show that the IDR including a Ser rich region (SRR) of the transcription factor Ets1, just before the DNA-binding core domain, undergoes multi-modal interactions when the SRR is not phosphorylated. In the phosphorylated state, the SRR forms a few specific complex structures with the Ets1 core, covering the recognition helix in the core and drastically reducing the DNA binding affinities as the auto-inhibitory state. The binding kinetics of mutated Ets1 indicates that aromatic residues in the SRR can be substituted with other hydrophobic residues for the interactions with the Ets1 core.
Figure 1
To learn more about this research, please view the full research report entitled " Phosphorylation of an intrinsically disordered region of Ets1 shifts a multi-modal interaction ensemble to an auto-inhibitory state " at this page of Nucleic Acids Research .
Related link
