Atomic resolution of muscle contraction
Osaka University researchers capture atomic images of muscle molecules in action, giving possibility of new nanomachines
At the molecular level, muscle contraction is defined by myosin molecules pulling actin filaments. New electron cryomicroscopy images with unprecedented resolution taken by researchers at Osaka University reveal unexpectedly large conformational changes in the myosin molecule during the pull. These findings, which can be seen in Nature Communications , provide new insights into how myosin generates force and a paradigm for the construction of nanomachines.
To biophysicists like Keiichi Namba, professor at Osaka University, the ability of tiny molecules to generate large amounts of force seen in muscle make myosin an engineering marvel.
"Myosin and actin are nanomachines that convert the chemical energy of ATP hydrolysis into mechanical work" he says.
Myosin converts this energy by hydrolyzing ATP molecules into movement along an actin filament. The hydrolysis involves a number of conformational changes in myosin. These changes have been imaged using electron microscopy, but, notes Takashii Fujii, specially appointed assistant professor, "there were no atomic images of ATP hydrolysis when myosin is interacting with actin," which would more accurately represent the changes myosin takes during muscle contraction.
Indeed, Fujii and Namba used electron cryomicroscopy to acquire images at 5.2 Å resolution to show a previously unobserved conformational change in the myosin molecule when it interacts with actin. The two scientists hypothesized that this conformation could explain why muscle myosin has far faster kinetics than other myosin in the body.
The images also give clues into how myosin moves along actin. The binding of ATP forces a rotation within the myosin molecule that reduces the number of bonds between it and actin. This weakly bound structure with a highly asymmetric bond distribution allows myosin to detach from actin and rebind preferentially in one direction, thus pulling the actin filament. Detachment toward the opposite direction would require a larger number of bonds to be broken at once, preventing detachment. This picture provides a new perspective on how myosin functions.
"This is a very unique image, because the weak binding state is unstable and it lifetime is short," noted Namba. He believes that this conformational state could be the reason why experiments on ATP hydrolysis by myosin without actin do not explain muscle contraction well.
"There is a structural asymmetry in the system", he said. "This could explain why myosin moves over much longer distance per ATP hydrolysis than expected."
Besides providing new molecular details on muscle contraction, Namba expects the structural information gained by this work could be used to make artificial nanomachines. "We are studying nature's nanomachines to build man-made ones," he said.
Abstract
Muscle contraction is driven by cyclic association and dissociation of myosin head of the thick filament with thin actin filament coupled with ATP binding and hydrolysis by myosin. However, because of the absence of actomyosin rigour structure at high resolution, it still remains unclear how the strong binding of myosin to actin filament triggers the release of hydrolysis products and how ATP binding causes their dissociation. Here we report the structure of mammalian skeletal muscle actomyosin rigour complex at 5.2 Å resolution by electron cryomicroscopy. Comparison with the structures of myosin in various states shows a distinctly large conformational change, providing insights into the ATPase-coupled reaction cycle of actomyosin. Based on our observations, we hypothesize that asymmetric binding along the actin filament could function as a Brownian ratchet by favouring directionally biased thermal motions of myosin and actin.
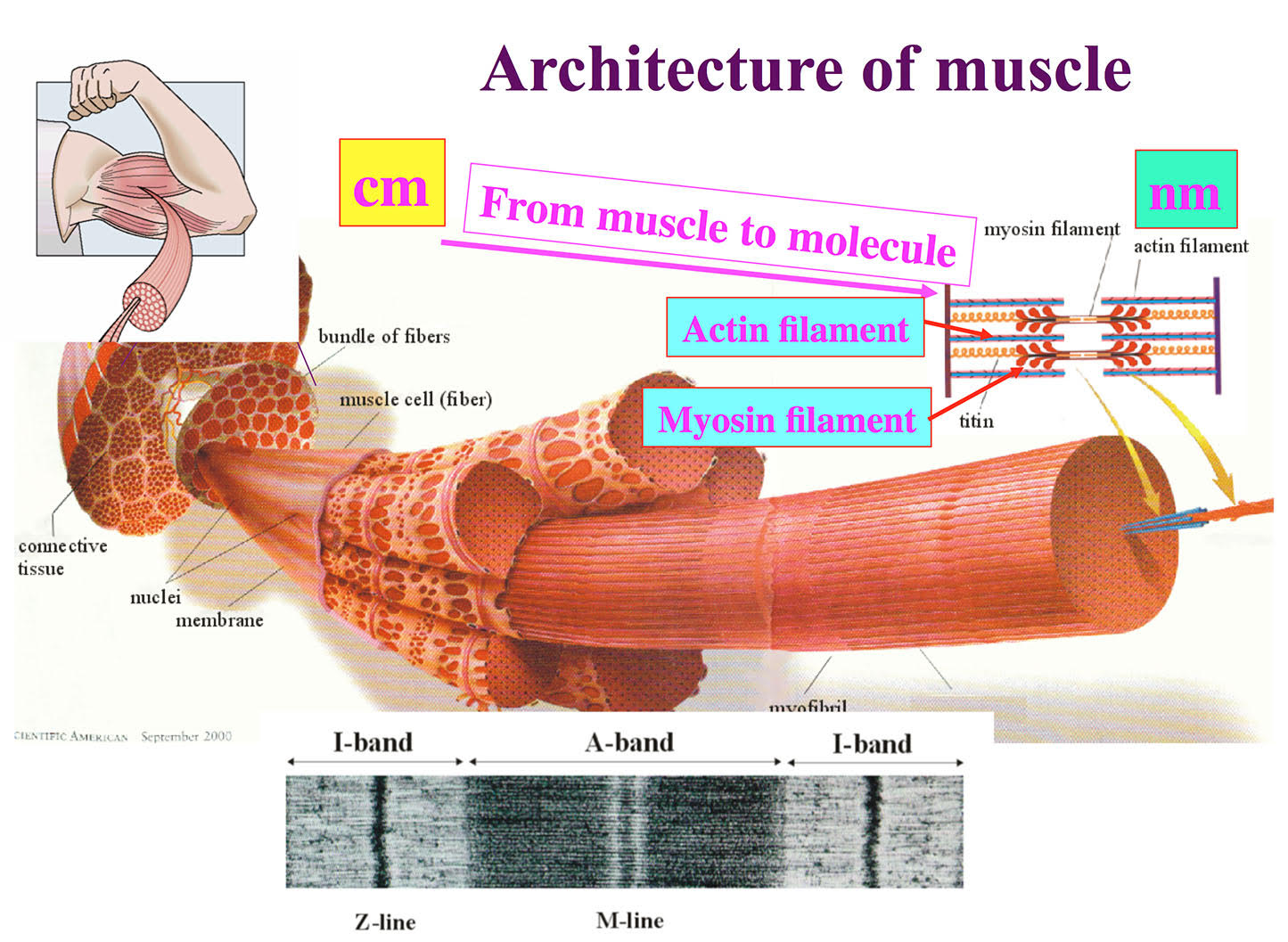
Figure 1. Architecture of skeletal muscle formed by regular arrays of myosin and actin filaments.
Figure 2. Structures of the actomyosin complex in the strong (upper panel) and weak (lower panel) binding states
Figure 3. The asymmetry in the weak binding structure of myosin head and actin filament that biases the direction of their dissociation by thermal fluctuation in one direction
To learn more about this research, please view the full research report entitled “ Structure of actomyosinrigor complex at 5.2-Å resolution and insights into the ATPase cycle mechanism ” at this page of the Nature Communications website.
Related link
