Clarification of mechanism behind efflux-mediated multidrug resistance
Under the leadership of NISHINO Kunihiko , Associate Professor, Institute of Scientific and Industrial Research, Osaka University, a group of researchers succeeded in determining the crystal structure of repressor protein RamR in the recognition of antibacterial compounds in multidrug resistance in salmonella bacteria.
Salmonella bacteria is the pathogen behind one of the most common types of food poisoning. Large scale food poisoning incidents from salmonella are not an unusual occurrence. Multiple drug resistance in a variety of pathogens has become an increasingly big problem.
In bacteria, multidrug efflux system gene expression is frequently controlled by activator protein RamA and repressor protein RamR. Pathogenic bacteria such as salmonella possess RamA, an activator protein for AcrAB. They also have RamR, a transcriptional repressor of the gene-encoding RamA protein that controls the expression of the multidrug efflux system genes.
In antibacterial substance-free conditions, RamR proteins bind to DNA and represss the expression of multidrug efflux system genes acrAB's activator RamA. However, when bacateria are exposed to antibacterial compounds, RamR proteins recognize these compounds, reducing suppression of RamA and thereby promoting multidrug efflux.
This group's clarification of the mechanism for antibiotic resistance in pathogenic bacteria is expected to help researchers gain a better understanding of transcriptional regulation mediated by RamR and its role in multidrug resistance in several enterobacterial pathogens.
Abstract
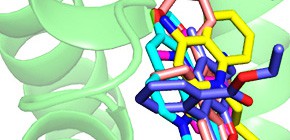
RamR is a transcriptional repressor of the gene-encoding RamA protein, which controls the expression of the multidrug efflux system genes acrAB-tolC. RamR is an important multidrug-resistance factor, however, its structure and the identity of the molecules to which it responds have been unknown. Here, we report the crystal structure of RamR in complex with multiple drugs, including berberine, crystal violet, dequalinium, ethidium bromide and rhodamine 6G. All compounds are found to interact with Phe155 of RamR, and each compound is surrounded by different amino acid residues. Binding of these compounds to RamR reduces its DNA-binding affinity, which results in the increased expression of ramA. Our results reveal significant flexibility in the substrate-recognition region of RamR, which regulates the bacterial efflux participating in multidrug resistance.

Figure 1

Figure 2

Figure 3
To learn more about this research, please read the full research report entitled "T he crystal structure of multidrug-resistance regulator RamR with multiple drugs " at this page of the Nature Communications website.
Related link :
