Clarification of Mechanism of Bacterial DNA Segregation
Under the leadership of FUJII Takashi (formerly, Researcher, Graduate School of Frontier Biosciences, Osaka University; currently, researcher at Riken), NAMBA Keiichi (Professor, Graduate School of Frontier Biosciences, Osaka University, and researcher at Riken), and Jan Löwe (Medical Research Council Laboratory of Molecular Biology, Cambridge University, U.K.), a group of researchers have clarified the mechanism of bacterial DNA segregation using methods of low-temperature electron microscopy and molecular imaging.
Abstract
" To ensure their stable inheritance by daughter cells during cell division, bacterial low copy-number plasmids make simple DNA segregating machines that use an elongating protein filament between sister plasmids. In the ParMRC system of Escherichia coli R1 plasmid, ParM, an actin-like protein, forms the spindle between ParRC complexes on sister plasmids. Using a combination of structural work and total internal reflection fluorescence microscopy, we show that ParRC bound and could accelerate growth at only one end of polar ParM filaments, mechanistically resembling eukaryotic formins. The architecture of ParM filaments enabled two ParRC-bound filaments to associate in an antiparallel orientation, forming a bipolar spindle. The spindle elongated as a bundle of at least two antiparallel filaments, thereby pushing two plasmid clusters toward the poles. "
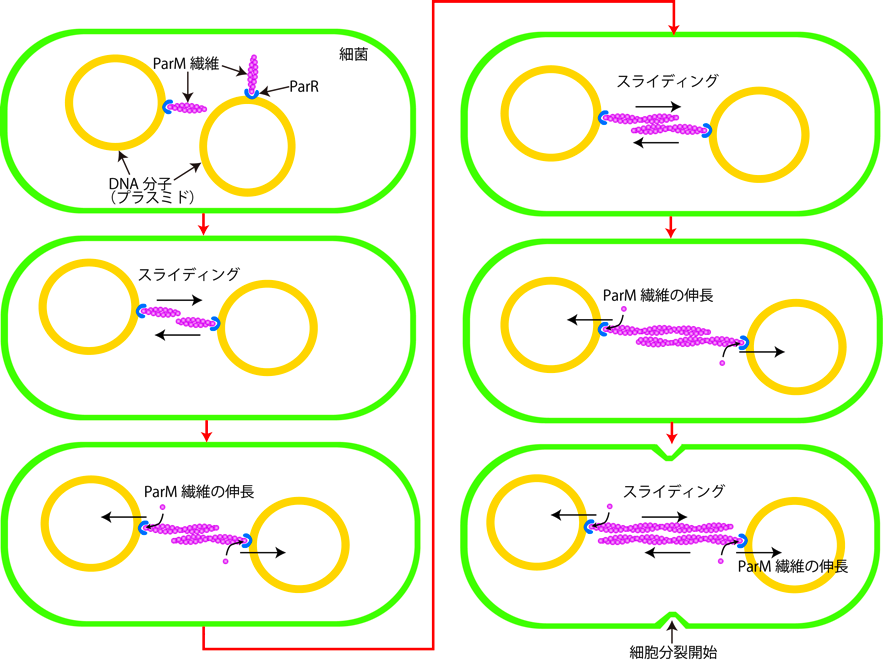
Figure 1
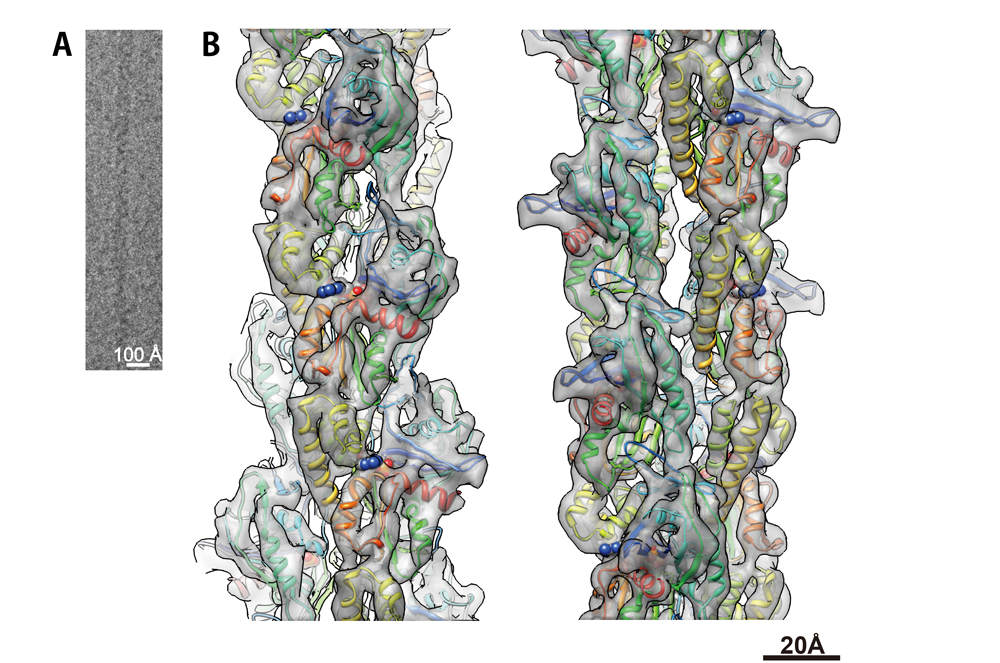
Figure 2
To learn more about this research, please read the full research report entitled " A Bipolar Spindle of Antiparallel ParM Filaments Drives Bacterial Plasmid Segregation " at this page of the Science website.
